6CFF
 
 | | Stimulator of Interferon Genes Human | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon genes protein | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
4A0M
 
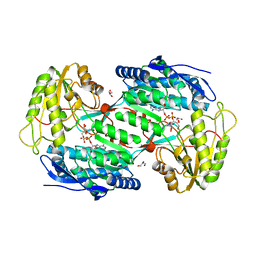 | | CRYSTAL STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE FROM SPINACH IN COMPLEX WITH NAD | | Descriptor: | BETAINE ALDEHYDE DEHYDROGENASE, CHLOROPLASTIC, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Diaz-Sanchez, A.G, Munoz-Clares, R.A. | | Deposit date: | 2011-09-09 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amino Acid Residues Critical for the Specificity for Betaine Aldehyde of the Plant Aldh10 Isoenzyme Involved in the Synthesis of Glycine Betaine.
Plant Physiol., 158, 2012
|
|
4A33
 
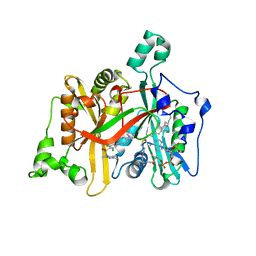 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 2,6-DICHLORO-4-(6-PIPERAZIN-1-YLPYRIDIN-3-YL)-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
4A4C
 
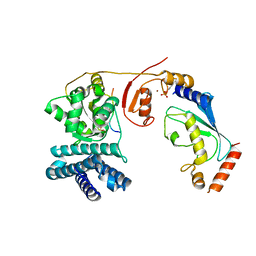 | | Structure of phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
8EY4
 
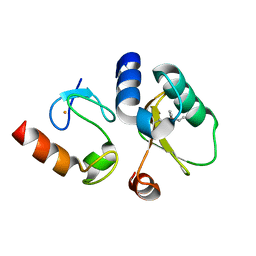 | | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37 | | Descriptor: | Cys_rich_CPCC domain-containing protein, FE (III) ION, PT-VENN domain-containing protein | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37
To Be Published
|
|
4A2S
 
 | | Structure of the engineered retro-aldolase RA95.5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
6R37
 
 | | T. brucei FPPS in complex with 2-(5-chlorobenzo[b]thiophen-3-yl)acetic acid | | Descriptor: | (5-chloro-1-benzothiophen-3-yl)acetic acid, DIMETHYL SULFOXIDE, Farnesyl pyrophosphate synthase | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-03-19 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Trypanosoma brucei FPPS by Fragment-based drug discovery
Thesis, 2019
|
|
6R3W
 
 | | M.tuberculosis nitrobindin with a water molecule coordinated to the heme iron atom | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, UPF0678 fatty acid-binding protein-like protein ERS007657_00996 | | Authors: | De Simone, G, di Masi, A, Polticelli, F, Pesce, A, Nardini, M, Bolognesi, M, Ciaccio, C, Coletta, M, Turilli, E.S, Fasano, M, Tognaccini, L, Smulevich, G, Abbruzzetti, S, Viappiani, C, Bruno, S, Ascenzi, P. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mycobacterial and Human Nitrobindins: Structure and Function.
Antioxid.Redox Signal., 33, 2020
|
|
8EY3
 
 | | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37 | | Descriptor: | Cys_rich_CPCC domain-containing protein, FE (III) ION, SODIUM ION | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37
To Be Published
|
|
4B4G
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
6RBL
 
 | | TETR(D) E147A MUTANT IN COMPLEX WITH DOXYCYCLINE AND MAGNESIUM | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hinrichs, W, Palm, G.J, Berndt, L, Girbardt, B. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
4B3Q
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, P51 RT, PRIMER DNA, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4ARW
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-04-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B08
 
 | | Yeast DNA polymerase alpha, Selenomethionine protein | | Descriptor: | DNA POLYMERASE ALPHA CATALYTIC SUBUNIT A | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-27 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Mechanism for Priming DNA Synthesis by Yeast DNA Polymerase Alpha
Elife, 2, 2013
|
|
4AVR
 
 | | Crystal structure of the hypothetical protein Pa4485 from Pseudomonas aeruginosa | | Descriptor: | PA4485 | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Alphey, M.S, Naismith, J.H. | | Deposit date: | 2012-05-29 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6R3Y
 
 | | M.tuberculosis nitrobindin with a cyanide molecule coordinated to the heme iron atom | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, UPF0678 fatty acid-binding protein-like protein ERS007657_00996 | | Authors: | De Simone, G, di Masi, A, Polticelli, F, Pesce, A, Nardini, M, Bolognesi, M, Ciaccio, C, Coletta, M, Turilli, E.S, Fasano, M, Tognaccini, L, Smulevich, G, Abbruzzetti, S, Viappiani, C, Bruno, S, Ascenzi, P. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mycobacterial and Human Nitrobindins: Structure and Function.
Antioxid.Redox Signal., 33, 2020
|
|
4B24
 
 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*AP*GP*GP*TP*AP*GP)-3', 5'-D(*GP*CP*TP*AP*CP*3DRP*TP*AP*TP*CP*GP)-3', PROBABLE DNA-3-METHYLADENINE GLYCOSYLASE 2 | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
4B2D
 
 | | human PKM2 with L-serine and FBP bound. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVATE KINASE ISOZYMES M1/M2, ... | | Authors: | Chaneton, B, Hillmann, P, Zheng, L, Martin, A.C.L, Maddocks, O.D.K, Chokkathukalam, A, Coyle, J.E, Jankevics, A, Holding, F.P, Vousden, K.H, Frezza, C, O'Reilly, M, Gottlieb, E. | | Deposit date: | 2012-07-13 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serine is a natural ligand and allosteric activator of pyruvate kinase M2.
Nature, 491, 2012
|
|
6R4S
 
 | |
4B6O
 
 | | Structure of Mycobacterium tuberculosis Type II Dehydroquinase inhibited by (2S)-2-(4-methoxy)benzyl-3-dehydroquinic acid | | Descriptor: | (1R,2S,4S,5R)-2-(4-methoxyphenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lence, E, Tizon, L, Peon, A, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic basis of the inhibition of type II dehydroquinase by (2S)- and (2R)-2-benzyl-3-dehydroquinic acids.
ACS Chem. Biol., 8, 2013
|
|
4B5E
 
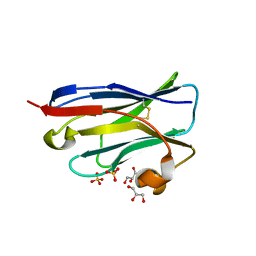 | | Crystal Structure of an amyloid-beta binding single chain antibody PS2-8 | | Descriptor: | GLYCEROL, PS2-8, SULFATE ION | | Authors: | Beringer, D.X, Dorresteijn, B, Rutten, L, Wienk, H, el Khattabi, M, Kroon-Batenburg, L.M.J, Verrips, C.T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Amyloid-Beta Binding Single Chain Antibody Ps2-8
To be Published
|
|
4A93
 
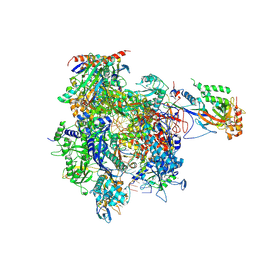 | | RNA Polymerase II elongation complex containing a CPD Lesion | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*T*TTP*TP*TP*CP*C BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP*AP)-3', ... | | Authors: | Walmacq, C, Cheung, A.C.M, Kireeva, M.L, Lubkowska, L, Ye, C, Gotte, D, Strathern, J.N, Carell, T, Cramer, P, Kashlev, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Translesion Transcription by RNA Polymerase II and its Role in Cellular Resistance to DNA Damage.
Mol.Cell, 46, 2012
|
|
8FBD
 
 | | Crystal structure of OrfX1-OrfX3 complex from Clostridium botulinum E1 | | Descriptor: | ACETATE ION, Neurotoxin complex component Orf-X1, Neurotoxin complex component Orf-X3 | | Authors: | Liu, S, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
4A49
 
 | | Structure of phosphoTyr371-c-Cbl-UbcH5B complex | | Descriptor: | E3 ubiquitin-protein ligase CBL, POTASSIUM ION, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-07 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl.
Nat. Struct. Mol. Biol., 19, 2012
|
|
8FBF
 
 | | Crystal structure of OrfX2 from Clostridium botulinum E1 | | Descriptor: | Neurotoxin complex component Orf-X2, SULFATE ION | | Authors: | Lam, K.H, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
