5Z7W
 
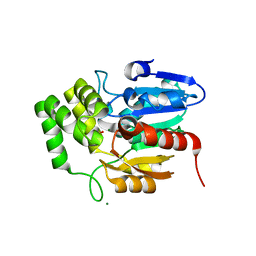 | | Crystal structure of Striga hermonthica HTL1 (ShHTL1) | | Descriptor: | GLYCEROL, Hyposensitive to light 1, MAGNESIUM ION, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
6AI6
 
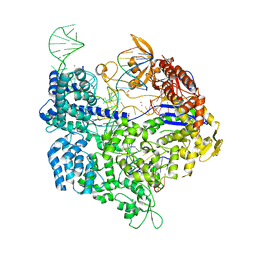 | | Crystal structure of SpCas9-NG | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), ... | | Authors: | Nishimasu, H, Hirano, S, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineered CRISPR-Cas9 nuclease with expanded targeting space
Science, 361, 2018
|
|
5X7X
 
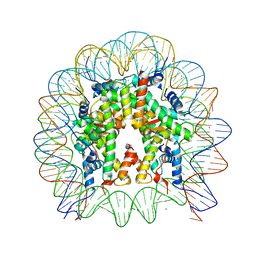 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.184 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
7YV1
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor LUNA18 and KA30L Fab | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, KA30L Fab H-chain, ... | | Authors: | Irie, M, Fukami, T.A, Matsuo, A, Saka, K, Nishimura, M, Saito, H, Torizawa, T, Tanada, M, Ohta, A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
6A78
 
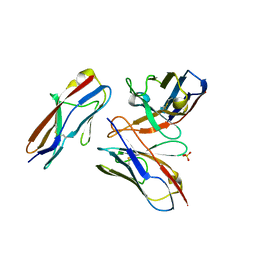 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the scFv fragment of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain and linker region of the anti-human Robo1 antibody B5209B scFv, Light chain region of the anti-human Robo1 antibody B5209B scFv, Roundabout homolog 1, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
8JYM
 
 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYK
 
 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYP
 
 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYN
 
 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYO
 
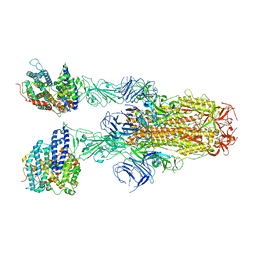 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
7YUZ
 
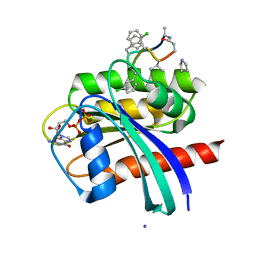 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP8784 | | Descriptor: | AP8784, GUANOSINE-5'-DIPHOSPHATE, IODIDE ION, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
7TLN
 
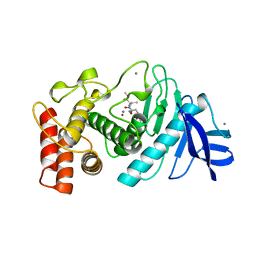 | | STRUCTURAL ANALYSIS OF THE INHIBITION OF THERMOLYSIN BY AN ACTIVE-SITE-DIRECTED IRREVERSIBLE INHIBITOR | | Descriptor: | 2-(ACETYL-HYDROXY-AMINO)-4-METHYL-PENTANOIC ACID METHYL ESTER, CALCIUM ION, THERMOLYSIN, ... | | Authors: | Matthews, B.W, Holmes, M.A, Tronrud, D.E. | | Deposit date: | 1983-01-27 | | Release date: | 1983-03-09 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the inhibition of thermolysin by an active-site-directed irreversible inhibitor.
Biochemistry, 22, 1983
|
|
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Suzuki, T, Kimura, K, Hashiguchi, T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
6JLP
 
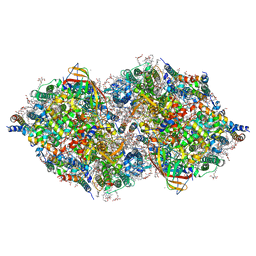 | | XFEL structure of cyanobacterial photosystem II (3F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLM
 
 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLK
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6P4V
 
 | |
6JLO
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6PGR
 
 | |
6JLL
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
5WQJ
 
 | |
6JLJ
 
 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
5WQK
 
 | |
6JLN
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6GF6
 
 | |
