1XBF
 
 | | X-RAY STRUCTURE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET CAR10 FROM C. ACETOBUTYLICUM | | Descriptor: | Clostridium acetobutylicum Q97KL0, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S, Yong, W, Acton, T, Ho, C.-K, Conover, K, Cooper, B, Ciano, M, Xiao, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-RAY STRUCTURE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET CAR10 FROM C. ACETOBUTYLICUM
To be published
|
|
4ADN
 
 | | Fusidic acid resistance protein FusB | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FAR1, ... | | Authors: | Guo, X, Peisker, K, Backbro, K, Chen, Y, Kiran, R.K, Sanyal, S, Selmer, M. | | Deposit date: | 2011-12-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of Fusb: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling
Open Biol., 2, 2012
|
|
1ZNP
 
 | | X-Ray Crystal Structure of Protein Q8U9W0 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR55. | | Descriptor: | hypothetical protein Atu3615 | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S.M, Xiao, R, Ma, L.-C, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the hypothetical protein Q8U9W0 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium target AtR55.
To be Published
|
|
4BV5
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 4-(aminomethyl)-N-(benzenesulfonyl)cyclohexanecarboxamide, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
3V36
 
 | | Aldose reductase complexed with glceraldehyde | | Descriptor: | Aldose reductase, D-Glyceraldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | Deposit date: | 2011-12-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
4BV7
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVD
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 5-chloro-2-fluoro-N-[1-(4-piperidyl)pyrazol-4-yl]benzenesulfonamide, APOLIPOPROTEIN(A), CHLORIDE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
1ZBR
 
 | | Crystal Structure of the Putative Arginine Deiminase from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR3 | | Descriptor: | conserved hypothetical protein | | Authors: | Forouhar, F, Chen, Y, Kuzin, A, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Putative Arginine Deiminase from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR3
To be Published
|
|
4BVC
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-PIPERIDYL)-N-[2-(TRIFLUOROMETHOXY)PHENYL]SULFONYL-PROPANAMIDE, APOLIPOPROTEIN(A), CHLORIDE ION, ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
3ZZT
 
 | |
1XW8
 
 | | X-ray structure of putative lactam utilization protein YBGL. Northeast Structural Genomics Consortium target ET90. | | Descriptor: | UPF0271 protein ybgL | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S.M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of putative lactam utilization protein YBGL.
Northeast Structural Genomics Consortium target ET90.
To be Published
|
|
4BVW
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1,2,3,4-tetrahydroisoquinoline-6-carboxylic acid, 1,2-ETHANEDIOL, APOLIPOPROTEIN(A), ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
2BX9
 
 | | Crystal structure of B.subtilis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | TRYPTOPHAN RNA-BINDING ATTENUATOR PROTEIN-INHIBITORY PROTEIN, ZINC ION | | Authors: | Shevtsov, M.B, Chen, Y, Gollnick, P, Antson, A.A. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Anti-Trap Protein, an Antagonist of Trap/RNA Interaction.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4BVV
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, APOLIPOPROTEIN(A), SULFATE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
1XME
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase from Thermus thermophilus | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide I, Cytochrome c oxidase polypeptide II, ... | | Authors: | Hunsicker-Wang, L.M, Pacoma, R.L, Chen, Y, Fee, J.A, Stout, C.D. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel cryoprotection scheme for enhancing the diffraction of crystals of recombinant cytochrome ba3 oxidase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XTO
 
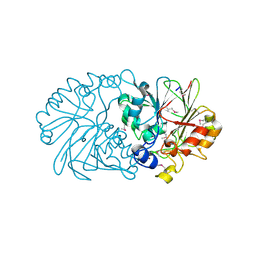 | | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6 | | Descriptor: | Coenzyme PQQ synthesis protein B, ZINC ION | | Authors: | Forouhar, F, Chen, Y, Kuzin, A, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-22 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6
To be Published
|
|
2LAS
 
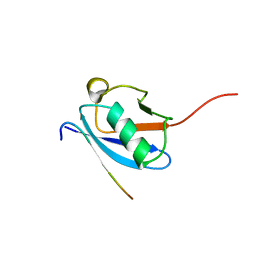 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
3ZNC
 
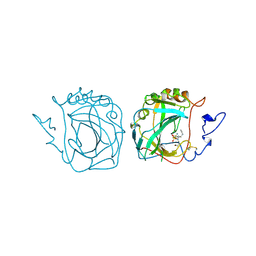 | | MURINE CARBONIC ANHYDRASE IV COMPLEXED WITH BRINZOLAMIDE | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE IV, ZINC ION | | Authors: | Stams, T, Chen, Y, Christianson, D.W. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination.
Protein Sci., 7, 1998
|
|
3ZTE
 
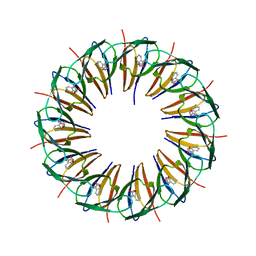 | |
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
2AJA
 
 | | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR21. | | Descriptor: | ankyrin repeat family protein | | Authors: | Kuzin, A.P, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, C, Kellie, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila.
To be Published
|
|
2B6E
 
 | | X-Ray Crystal Structure of Protein HI1161 from Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR63. | | Descriptor: | ACETIC ACID, Hypothetical UPF0152 protein HI1161 | | Authors: | Kuzin, A.P, Benach, J, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Kellie, R, Cunningham, K.E, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-01 | | Release date: | 2005-10-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray structure of the different crystal form of the hypothetical UPF0152 protein HI1161: NESG target IR63
TO BE PUBLISHED
|
|
2KDM
 
 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | DESIGNED PROTEIN | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From the Cover: A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2LHC
 
 | | Ga98 solution structure | | Descriptor: | Ga98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
6LRR
 
 | | Cryo-EM structure of RuBisCO-Raf1 from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
