7YMN
 
 | | Cryo-EM structure of in vitro PHF fibril | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Li, X, Liu, C. | | Deposit date: | 2022-07-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Subtle change of fibrillation condition leads to substantial alteration of recombinant Tau fibril structure.
Iscience, 25, 2022
|
|
7YPG
 
 | |
4R8T
 
 | | Structure of JEV protease | | Descriptor: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | Authors: | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
8JEX
 
 | | Cryo-EM structure of alpha-synuclein gS87 fibril | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-synuclein, unverified amino acid chain | | Authors: | Xia, W.C, Sun, Y.P, Liu, C. | | Deposit date: | 2023-05-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Phosphorylation and O-GlcNAcylation at the same alpha-synuclein site generate distinct fibril structures.
Nat Commun, 15, 2024
|
|
8JEY
 
 | |
8IXZ
 
 | |
8IY1
 
 | | Structure of Acb2 complexed with cAAG | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, p26 | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8IY0
 
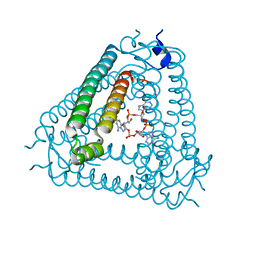 | | Structure of Acb2 complexed with cAAA | | Descriptor: | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, p26 | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8IY2
 
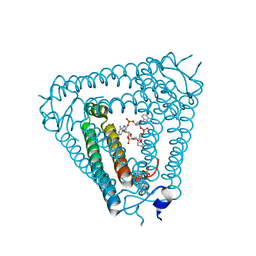 | | Structure of Acb2 complexed with 3',3'-cGAMP and cAAA | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-3'-MONOPHOSPHATE, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, ... | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8J8O
 
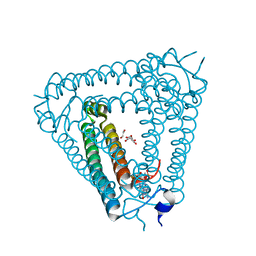 | | Structure of Acb2 complexed with 2',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, cGAMP, ... | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4R8U
 
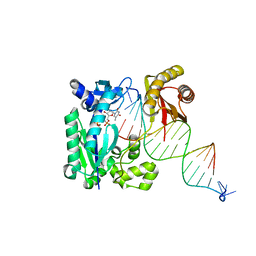 | | S-SAD structure of DINB-DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA, DNA polymerase IV, ... | | Authors: | Kottur, J, Nair, D.T, Weinert, T, Oligeric, V, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination
Nat.Methods, 12, 2015
|
|
8IUM
 
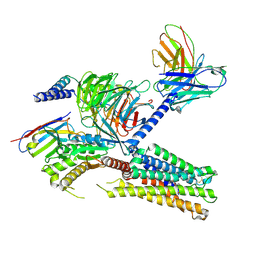 | | Cryo-EM structure of the tafluprost acid-bound human PTGFR-Gq complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E})-3,3-bis(fluoranyl)-4-phenoxy-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUK
 
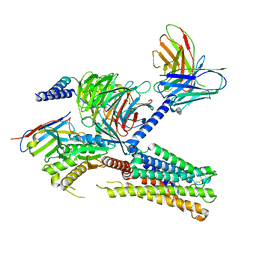 | | Cryo-EM structure of the PGF2-alpha-bound human PTGFR-Gq complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUL
 
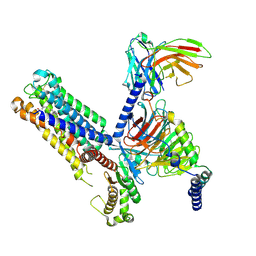 | | Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex | | Descriptor: | Antibody fragment scFv16, G subunit alpha (q), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
4PGO
 
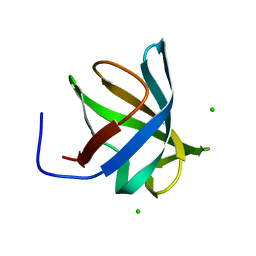 | | Crystal structure of hypothetical protein PF0907 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4PII
 
 | | Crystal structure of hypothetical protein PF0907 from pyrococcus furiosus solved by sulfur SAD using Swiss light source data | | Descriptor: | CHLORIDE ION, IMIDAZOLE, N-glycosylase/DNA lyase | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4TNO
 
 | | Hypothetical protein PF1117 from Pyrococcus Furiosus: Structure solved by sulfur-SAD using Swiss Light Source Data | | Descriptor: | CHLORIDE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
7X83
 
 | | Cryo-EM structure of the TMEM106B fibril from normal elder | | Descriptor: | Transmembrane protein 106B | | Authors: | Xia, W.C, Zhao, Q.Y, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
7X84
 
 | | Cryo-EM structure of the TMEM106B fibril from Parkinson's disease dementia | | Descriptor: | Transmembrane protein 106B | | Authors: | Zhao, Q.Y, Xia, W.C, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
6L1T
 
 | | Cryo-EM structure of phosphorylated Tyr39 a-synuclein amyloid fibril | | Descriptor: | Alpha-synuclein | | Authors: | Liu, C, Li, Y.M, Zhao, K, Lim, Y.J, Liu, Z.Y. | | Deposit date: | 2019-09-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Parkinson's disease-related phosphorylation at Tyr39 rearranges alpha-synuclein amyloid fibril structure revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
6X2B
 
 | |
6X2C
 
 | |
6X5Y
 
 | | IDO1 in complex with compound 4 | | Descriptor: | 4-fluoro-N-{1-[5-(2-methylpyrimidin-4-yl)-5,6,7,8-tetrahydro-1,5-naphthyridin-2-yl]cyclopropyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Utilization of MetID and Structural Data to Guide Placement of Spiro and Fused Cyclopropyl Groups for the Synthesis of Low Dose IDO1 Inhibitors
To Be Published
|
|
