4NFB
 
 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7WBQ
 
 | |
7WBP
 
 | |
6J2A
 
 | | The structure of HLA-A*3003/NP44 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, NP44 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J9O
 
 | |
6J29
 
 | | The structure of HLA-A*3003/MTB | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, MTB | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
7BSD
 
 | |
7BSC
 
 | | Complex structure of 1G5.3 Fab bound to DENV2 NS1c | | Descriptor: | 1G5.3 Fab Heavy Chain, 1G5.3 Fab Light Chain, Non-structural protein 1 | | Authors: | Song, H, Qi, J, Gao, F.G. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A broadly protective antibody that targets the flavivirus NS1 protein.
Science, 371, 2021
|
|
7C9V
 
 | | E30 F-particle in complex with FcRn | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, MYRISTIC ACID, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9S
 
 | | Echovirus 30 F-particle | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Wang, K, Sun, Y, Zhu, L, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C6U
 
 | | Crystal structure of SARS-CoV-2 complexed with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Fu, L, Feng, Y, Qi, J, Gao, F.G. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
7C9W
 
 | | E30 F-particle in complex with CD55 | | Descriptor: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9U
 
 | | Echovirus 30 E-particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
6JO8
 
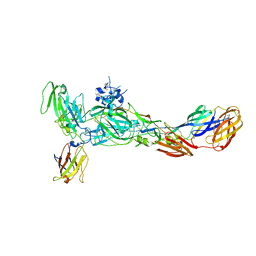 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
4ZUU
 
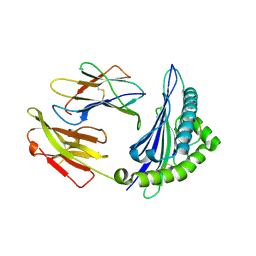 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-CF9 | | Descriptor: | Beta-2-microglobulin, CYS-THR-SER-GLU-GLU-MET-ASN-ALA-PHE, Classical MHC class I antigen | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
4ZUV
 
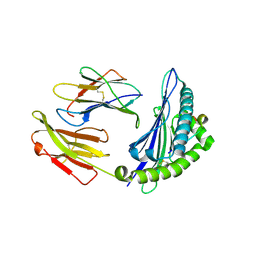 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Env-RW12 | | Descriptor: | ARG-VAL-GLU-ASP-VAL-THR-ASN-THR-ALA-GLU-TYR-TRP, Beta-2-microglobulin, Classical MHC class I antigen | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
4ZUT
 
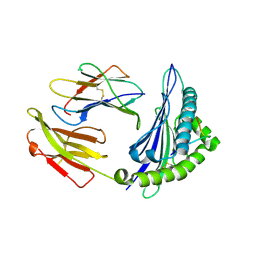 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-GW12 | | Descriptor: | Beta-2-microglobulin, Classical MHC class I antigen, GLY-SER-GLN-LYS-LEU-THR-THR-GLY-ASN-CYS-ASN-TRP | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
4ZUS
 
 | | Crystal structure of Equine MHC I(Eqca-N*00602) in complexed with equine infectious anaemia virus (EIAV) derived peptide REV-QW11 | | Descriptor: | Beta-2-microglobulin, Classical MHC class I antigen, GLN-ALA-GLU-VAL-LEU-GLN-GLU-ARG-LEU-GLU-TRP | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
4ZUW
 
 | | Crystal structure of Equine MHC I(Eqca-N*00601) in complexed with equine infectious anaemia virus (EIAV) derived peptide Gag-GW12 | | Descriptor: | Beta-2-microglobulin, Classical MHC class I antigen, GLY-SER-GLN-LYS-LEU-THR-THR-GLY-ASN-CYS-ASN-TRP | | Authors: | Yao, S, Liu, J, Qi, J, Chen, R, Zhang, N, Liu, Y, Xia, C. | | Deposit date: | 2015-05-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
J Immunol., 196, 2016
|
|
9IM1
 
 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA in intermediate state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
9IM0
 
 | |
9ILY
 
 | |
9ILZ
 
 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
9IM2
 
 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA in intermediate state 3 | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
5YMU
 
 | |
