7WKC
 
 | |
5EQ0
 
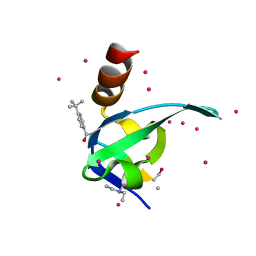 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EPK
 
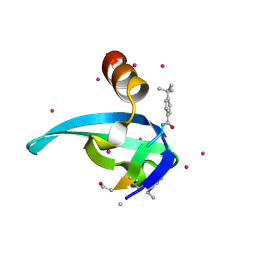 | | Crystal Structure of chromodomain of CBX2 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 2, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EPL
 
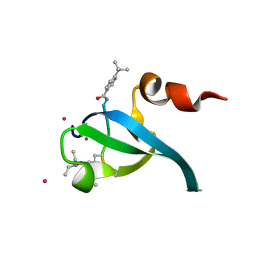 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EQP
 
 | | Crystal structure of choline kinase alpha-1 bound by 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline (compound 37) | | Descriptor: | 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5F3W
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | 27-MER DNA, DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex
Embo J., 35, 2016
|
|
5H06
 
 | | Crystal structure of AmyP in complex with maltose | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
5H1U
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 2 | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5VSF
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 17 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5H1T
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 1 | | Descriptor: | DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ZINC ION, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5VSE
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 17: N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
7CQQ
 
 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
5H05
 
 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
5VSD
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 13 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, GLYCEROL, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
7CQL
 
 | | Apo GmaS without ligand | | Descriptor: | Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
5H1D
 
 | | Crystal structure of C-terminal of RhoGDI2 | | Descriptor: | Rho GDP-dissociation inhibitor 2 | | Authors: | Liu, J. | | Deposit date: | 2016-10-08 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | NMR characterization of weak interactions between RhoGDI2 and fragment screening hits.
Biochim. Biophys. Acta, 1861, 2017
|
|
7XS4
 
 | | Crystal structure of URT1 in complex with AAAU RNA | | Descriptor: | RNA (5'-R(*AP*AP*AP*U)-3'), UTP:RNA uridylyltransferase 1 | | Authors: | Hu, Q, Zhu, L.R, lv, M.Q, Gong, Q.G. | | Deposit date: | 2022-05-12 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Molecular mechanism underlying the di-uridylation activity of Arabidopsis TUTase URT1.
Nucleic Acids Res., 50, 2022
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
5GMQ
 
 | | Structure of MERS-CoV RBD in complex with a fully human antibody MCA1 | | Descriptor: | 1,2-ETHANEDIOL, MCA1 heavy chain, MCA1 light chain, ... | | Authors: | Chen, C, Wang, J.M, Zou, T.T, Gao, X.P, Cui, S, Jin, Q. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset.
J. Infect. Dis., 215, 2017
|
|
7CM9
 
 | | DMSP lyase DddX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DMSP lyase, SULFATE ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-07-25 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.
Elife, 10, 2021
|
|
5H1V
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 6 | | Descriptor: | 2-Hydrazino-1,3-benzothiazole-6-carbohydrazide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
5VSC
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 13 | | Descriptor: | 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
2MXP
 
 | |
5GH9
 
 | | Crystal structure of CBP Bromodomain with H3K56ac peptide | | Descriptor: | CREB-binding protein, Histone H3 | | Authors: | Xu, L. | | Deposit date: | 2016-06-19 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insight into CBP bromodomain-mediated recognition of acetylated histone H3K56ac
FEBS J., 2017
|
|
