5K7R
 
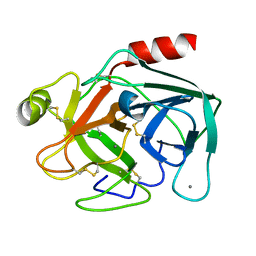 | | MicroED structure of trypsin at 1.7 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
9BAP
 
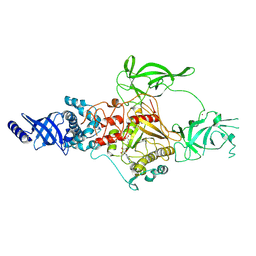 | | CryoEM structure of Apo-DIM2 | | Descriptor: | DNA (cytosine-5-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
6OI8
 
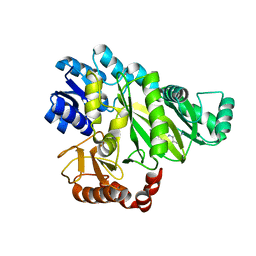 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with 7-((1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl)-6-(2-chloro-6-(pyridin-3-yl)phenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl]-6-[2-chloro-6-(pyridin-3-yl)phenyl]pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
9B9V
 
 | |
5JOX
 
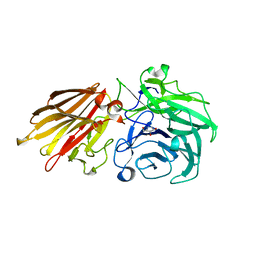 | | Bacteroides ovatus Xyloglucan PUL GH43A in complex with AraDNJ | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, Non-reducing end alpha-L-arabinofuranosidase BoGH43A | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
9B9R
 
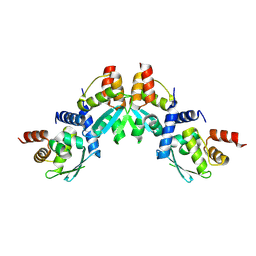 | |
9BAZ
 
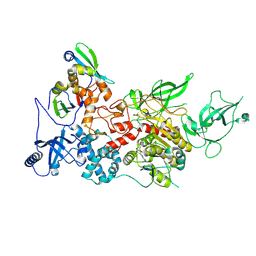 | | CryoEM structure of DIM2-HP1 complex | | Descriptor: | DNA (cytosine-5-)-methyltransferase, Heterochromatin protein one, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
6OSK
 
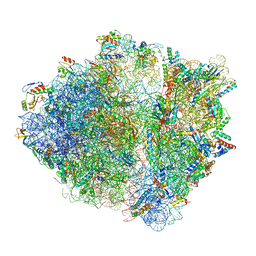 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OA8
 
 | | Superfolder Green Fluorescent Protein with 4-cyano-L-phenylalanine at the chromophore (position 66) | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, SODIUM ION, ... | | Authors: | Piacentini, J, Olenginski, G.M, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
6OAG
 
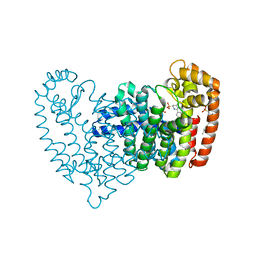 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02-82 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [(1S)-1-{[6-(3-chloro-4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Park, J, Berghuis, A.M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
9BAQ
 
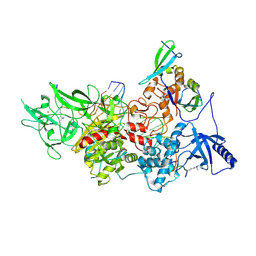 | | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*CP*T)-R(P*(PYO))-D(P*CP*TP*CP*CP*TP*CP*CP*TP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*TP*AP*GP*T)-3'), DNA (cytosine-5-)-methyltransferase, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
6OD7
 
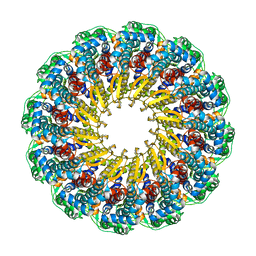 | | Herpes simplex virus type 1 (HSV-1) pUL6 portal protein, dodecameric complex | | Descriptor: | Portal protein | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
6OGK
 
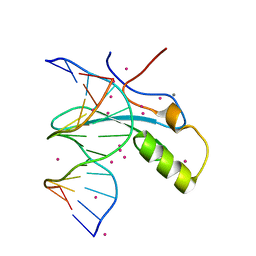 | | MeCP2 MBD in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
5K0X
 
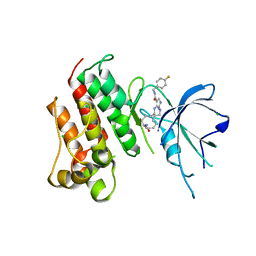 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
6OJH
 
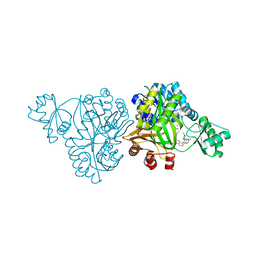 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine, ACETATE ION, Biotin carboxylase, ... | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine
To Be Published
|
|
6OL3
 
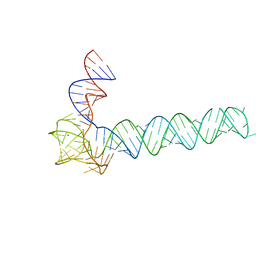 | | Crystal structure of an adenovirus virus-associated RNA | | Descriptor: | Adenovirus Virus-Associated (VA) RNA I apical and central domains, POTASSIUM ION | | Authors: | Hood, I.V, Gordon, J.M, Bou-Nader, C, Henderson, F.V, Bahmanjah, S, Zhang, J. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of an adenovirus virus-associated RNA.
Nat Commun, 10, 2019
|
|
6O57
 
 | |
5K8X
 
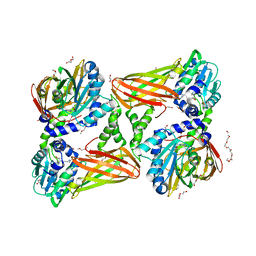 | | Crystal structure of mouse CARM1 in complex with inhibitor U3 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor U3
To Be Published
|
|
6O6P
 
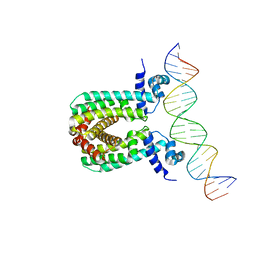 | | Structure of the regulator FasR from Mycobacterium tuberculosis in complex with DNA | | Descriptor: | DNA-forward, DNA-reverse, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.851 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
5K8W
 
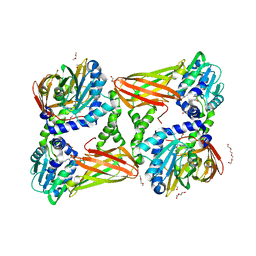 | | Crystal structure of mouse CARM1 in complex with inhibitor U2 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor U2
To Be Published
|
|
6OII
 
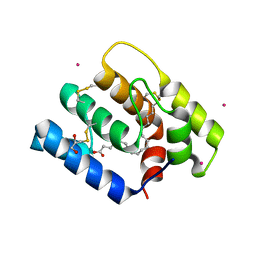 | |
6OC5
 
 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
5JOV
 
 | | Bacteroides ovatus Xyloglucan PUL GH31 with bound 5FIdoF | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-fluoro-alpha-L-idopyranose, ... | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
6IAN
 
 | | T. brucei IFT22/74/81 GTP-bound crystal structure | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Intraflagellar transport protein 74, Intraflagellar transport protein 81, ... | | Authors: | Wachter, S, Basquin, J, Lorentzen, E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Embo J., 38, 2019
|
|
6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
