5WQM
 
 | |
5X23
 
 | | Crystal structure of CYP2C9 genetic variant A477T (*30) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5X24
 
 | | Crystal structure of CYP2C9 genetic variant I359L (*3) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5WQP
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition II) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, PHOSPHATE ION, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WRW
 
 | | Structure of human apo-SRP72 | | Descriptor: | SULFATE ION, Signal recognition particle subunit SRP72 | | Authors: | Gao, Y, Chen, Z. | | Deposit date: | 2016-12-04 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
J Mol Cell Biol, 9, 2017
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y0I
 
 | | Solution structures of ASH1L PHD domain in complex with H3K4me2 peptide | | Descriptor: | ALA-ARG-THR-MLY-GLN-THR-ALA-ARG-LYS-SER-THR-GLY-GLY-LYS-ALA, Histone-lysine N-methyltransferase ASH1L, ZINC ION | | Authors: | Yu, M, Zeng, L. | | Deposit date: | 2022-06-05 | | Release date: | 2022-10-12 | | Method: | SOLUTION NMR | | Cite: | Structural insight into ASH1L PHD finger recognizing methylated histone H3K4 and promoting cell growth in prostate cancer.
Front Oncol, 12, 2022
|
|
6V8Z
 
 | |
6V8X
 
 | | VRC01 Bound BG505 F14 HIV-1 SOSIP Envelope Trimer Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-12-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Disruption of the HIV-1 Envelope allosteric network blocks CD4-induced rearrangements.
Nat Commun, 11, 2020
|
|
6M0J
 
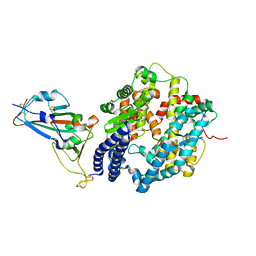 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
7RE9
 
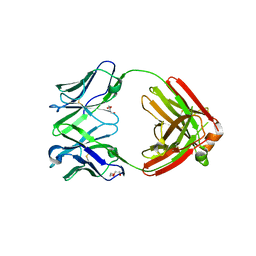 | | TCR mimic antibody (Fab fragment) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fab heavy chain, Fab light chain | | Authors: | Dasgupta, M, Baker, B.M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Sci Rep, 12, 2022
|
|
7RE8
 
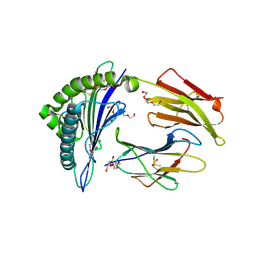 | |
7RE7
 
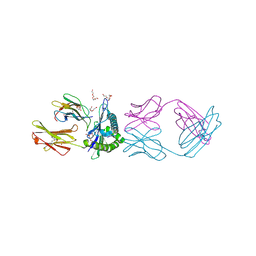 | | TCR mimic antibody (Fab fragment) in complex with AFP/HLA-A*02 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, FORMIC ACID, ... | | Authors: | Dasgupta, M, Baker, B.M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Sci Rep, 12, 2022
|
|
4R5P
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | Descriptor: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7CYB
 
 | | Saimiri boliviensis boliviensis galectin-13 with glycerol | | Descriptor: | GLYCEROL, Galectin | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
7CYA
 
 | | Saimiri boliviensis boliviensis galectin-13 with lactose | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
3KLI
 
 | |
3KLG
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to pre-translocation AZTMP-Terminated DNA (COMPLEX N) | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), Reverse transcriptase/ribonuclease H, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KLH
 
 | | Crystal structure of AZT-Resistant HIV-1 Reverse Transcriptase crosslinked to post-translocation AZTMP-Terminated DNA (COMPLEX P) | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6MEV
 
 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
3R4L
 
 | | Human very long half life Plasminogen Activator Inhibitor type-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasminogen activator inhibitor 1, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Zheng, H, Han, Q, Skrzypczak-Jankun, E, Jankun, J. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Remarkable extension of PAI-1 half-life surprisingly brings no changes to its structure.
Int.J.Mol.Med., 29, 2012
|
|
5IZ6
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
