1Q6S
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 9 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-((2R)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-METHYLQUINOLIN-8-YLPHOSPHONIC ACID, CHLORIDE ION, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1YF6
 
 | | Structure of a quintuple mutant of photosynthetic reaction center from rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Paddock, M.L, Chang, C, Xu, Q, Abresch, E.C, Axelrod, H.L. | | Deposit date: | 2004-12-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Quinone (Q(B)) Reduction by B-Branch Electron Transfer in Mutant Bacterial Reaction Centers from Rhodobacter sphaeroides: Quantum Efficiency and X-ray Structure.
Biochemistry, 44, 2005
|
|
6BBX
 
 | | Crystal structure of TnmS3 in complex with TNM C | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
1Q6J
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 2 | | Descriptor: | Protein-tyrosine phosphatase, non-receptor type 1, [4-(2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL](DIFLUORO)METHYLPHOSPHONIC ACID | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
2G7S
 
 | | The crystal structure of transcriptional regulator, TetR family, from Agrobacterium tumefaciens | | Descriptor: | transcriptional regulator, TetR family | | Authors: | Lunin, V.V, Chang, C, Xu, X, Gu, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of transcriptional regulator, TetR family, from Agrobacterium tumefaciens
To be Published
|
|
8FDS
 
 | | Ankyrin domain of SKD3 isoform 2 | | Descriptor: | Caseinolytic peptidase B protein homolog, FORMIC ACID | | Authors: | Lee, S, Tsai, F.T.F, Chang, C. | | Deposit date: | 2022-12-04 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of impaired disaggregase function in the oxidation-sensitive SKD3 mutant causing 3-methylglutaconic aciduria.
Nat Commun, 14, 2023
|
|
2ESN
 
 | | The crystal structure of probable transcriptional regulator PA0477 from Pseudomonas aeruginosa | | Descriptor: | probable transcriptional regulator | | Authors: | Lunin, V.V, Chang, C, Skarina, T, Gorodischenskaya, E, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of putative transcriptional regulator Pa0477 from Pseudomonas aeruginosa
To be Published
|
|
1Q6M
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 3 | | Descriptor: | Protein-tyrosine phosphatase, non-receptor type 1, {[2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-(3,4-DIFLUOROPHENYL)PROPANE-1,3-DIYL]BIS[4,1-PHENYLENE(DIFLUOROMETHYLENE)]}BIS(PHOSPHONIC ACID) | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6P
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 4'-((2S)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)-1,1'-BIPHENYL-3-YLPHOSPHONIC ACID, CHLORIDE ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
2A61
 
 | | The crystal structure of transcriptional regulator Tm0710 from Thermotoga maritima | | Descriptor: | transcriptional regulator Tm0710 | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Chang, C, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-01 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transcriptional regulator Tm0710 from Thermotoga maritima
To be Published
|
|
4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
1DGS
 
 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
1ECU
 
 | | SOLUTION STRUCTURE OF E2F BINDING DNA FRAGMENT GCGCGAAAC-T-GTTTCGCGC | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*AP*AP*AP*CP*TP*GP*TP*TP*TP*CP*GP*CP*GP*C)-3') | | Authors: | Wu, J.H, Chang, C, Pei, J.M, Xiao, Q, Shi, Y.Y. | | Deposit date: | 2000-01-26 | | Release date: | 2000-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of E2F binding DNA fragment GCGCGAAAC-T-GTTTCGCGC studied by Molecular Dynamics Simulation and Two Dimensional NMR experiment
to be published, 2000
|
|
4DGF
 
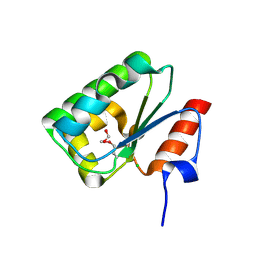 | | Structure of SulP Transporter STAS Domain from Wolinella Succinogenes Refined to 1.6 Angstrom Resolution | | Descriptor: | CHLORIDE ION, FORMIC ACID, SULFATE TRANSPORTER SULFATE TRANSPORTER FAMILY PROTEIN | | Authors: | Keller, J.P, Chang, C, Tesar, C, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Wolinella Succinogenes Refined to 1.6 Angstrom Resolution
To be Published
|
|
1VJS
 
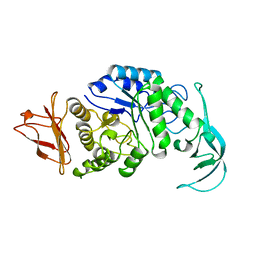 | |
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
2CFA
 
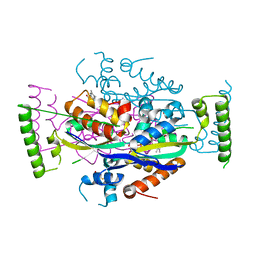 | | Structure of viral flavin-dependant thymidylate synthase ThyX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THYMIDYLATE SYNTHASE | | Authors: | Graziani, S, Bernauer, J, Skouloubris, S, Graille, M, Zhou, C.-Z, Marchand, C, Decottignies, P, van Tilbeurgh, H, Myllykallio, H, Liebl, U. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Mechanism and Structure of Viral Flavin-Dependent Thymidylate Synthase Thyx.
J.Biol.Chem., 281, 2006
|
|
3VCX
 
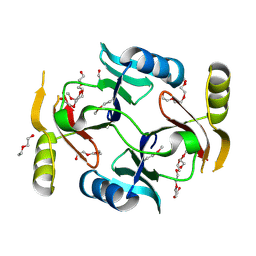 | | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009 | | Descriptor: | Glyoxalase/Bleomycin resistance protein/dioxygenase domain, TETRAETHYLENE GLYCOL | | Authors: | Stogios, P.J, Chang, C, Evdokimova, E, Egorova, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009
To be Published
|
|
1HYE
 
 | | CRYSTAL STRUCTURE OF THE MJ0490 GENE PRODUCT, THE FAMILY OF LACTATE/MALATE DEHYDROGENASE, DIMERIC STRUCTURE | | Descriptor: | L-LACTATE/MALATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lee, B.I, Chang, C, Cho, S.-J, Suh, S.W. | | Deposit date: | 2001-01-19 | | Release date: | 2001-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the MJ0490 gene product of the hyperthermophilic archaebacterium Methanococcus jannaschii, a novel member of the lactate/malate family of dehydrogenases.
J.Mol.Biol., 307, 2001
|
|
1HYG
 
 | | Crystal structure of MJ0490 gene product, the family of lactate/malate dehydrogenase | | Descriptor: | L-LACTATE/MALATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lee, B.I, Chang, C, Cho, S.-J, Suh, S.W. | | Deposit date: | 2001-01-19 | | Release date: | 2001-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the MJ0490 gene product of the hyperthermophilic archaebacterium Methanococcus jannaschii, a novel member of the lactate/malate family of dehydrogenases
J.Mol.Biol., 307, 2001
|
|
2XQB
 
 | | Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 | | Descriptor: | ANTI-IL-15 ANTIBODY, INTERLEUKIN 15, SULFATE ION | | Authors: | Lowe, D.C, Gerhardt, S, Ward, A, Hargreaves, D, Anderson, M, StGallay, S, Vousden, K, Ferraro, F, Pauptit, R.A, Cochrane, D, Pattison, D.V, Buchanan, C, Popovic, B, Finch, D.K, Wilkinson, T, Sleeman, M, Vaughan, T.J, Cruwys, S, Mallinder, P.R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
J.Mol.Biol., 406, 2011
|
|
4DGH
 
 | | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution | | Descriptor: | GLYCEROL, IODIDE ION, POTASSIUM ION, ... | | Authors: | Keller, J.P, Chang, C, Marshall, N, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution
To be Published
|
|
4DPD
 
 | | WILD TYPE PLASMODIUM FALCIPARUM DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE (PfDHFR-TS), DHF COMPLEX, NADP+, dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, DIHYDROFOLIC ACID, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DDR
 
 | | Human dihydrofolate reductase complexed with NADPH and P218 | | Descriptor: | 3-(2-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)propanoic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yuthavong, Y, Tarnchompoo, B, Vilaivan, T, Chitnumsub, P, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Thongphanchang, C, Taweechai, S, Vanichtanankul, J, Rattanajak, R, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DPH
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with P65 and NADPH | | Descriptor: | 2,4-diamino-6-methyl-5-[3-(2,4,5-trichlorophenoxy)propyloxy]pyrimidine, BETA-MERCAPTOETHANOL, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
