3QD7
 
 | | Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli | | Descriptor: | Uncharacterized protein ydaL | | Authors: | Gui, W.J, Qu, Q.H, Chen, Y.Y, Wang, M, Zhang, X.E, Bi, L.J, Jiang, T. | | Deposit date: | 2011-01-18 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli.
J.Struct.Biol., 174, 2011
|
|
7V35
 
 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7VBI
 
 | | Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
5OON
 
 | | Structure of Undecaprenyl-Pyrophosphate Phosphatase, BacA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Wang, M, Howe, N, Ghachi, M.E.I, Weichert, D, Kerff, F, Stansfeld, P, Touze, T, Caffrey, M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis.
Nat Commun, 9, 2018
|
|
4FIU
 
 | | The structure of hemagglutinin of H16 subtype influenza virus with V327G mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Wang, M, Qi, J, Gao, G.F. | | Deposit date: | 2012-06-11 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
5N6H
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
8W13
 
 | | Crystal structure of MYST acetyltransferase domain in complex with N-(1-(5-bromo-2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)-2-methoxybenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone acetyltransferase KAT8, ... | | Authors: | Chen, C, Dou, Y, Wang, M, Xu, C, Buesking, A. | | Deposit date: | 2024-02-15 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identification of triazolyl KAT6 inhibitors via a templated fragment approach.
Bioorg.Med.Chem.Lett., 113, 2024
|
|
2FHG
 
 | | Crystal Structure of Mycobacterial Tuberculosis Proteasome | | Descriptor: | 20S proteasome, alpha and beta subunits, proteasome, ... | | Authors: | Hu, G, Lin, G, Wang, M, Dick, L, Xu, R.M, Nathan, C, Li, H. | | Deposit date: | 2005-12-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate.
Mol.Microbiol., 59, 2006
|
|
5N6M
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
3QYT
 
 | | Diferric bound human serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Yang, N, Zhang, H, Wang, M, Hao, Q, Sun, H. | | Deposit date: | 2011-03-03 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe
Sci Rep, 2, 2012
|
|
3U5U
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3VTP
 
 | | HIV fusion inhibitor MT-C34 | | Descriptor: | Transmembrane protein gp41 | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2012-06-02 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The M-T hook structure is critical for design of HIV-1 fusion inhibitors.
J.Biol.Chem., 287, 2012
|
|
7CGC
 
 | |
3OOI
 
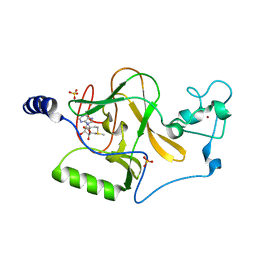 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Qiao, Q, Wang, M, Xu, R.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
2D41
 
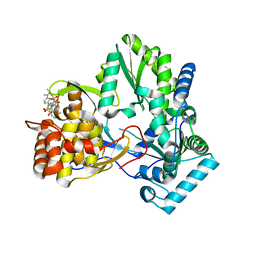 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor | | Descriptor: | 5'-ACETYL-4-{[(2,4-DIMETHYLPHENYL)SULFONYL]AMINO}-2,2'-BITHIOPHENE-5-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-05 | | Release date: | 2006-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3Z
 
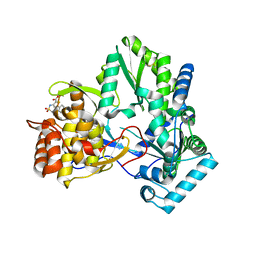 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
4H0W
 
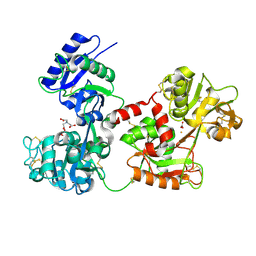 | | Bismuth bound human serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bismuth(III) ION, CARBONATE ION, ... | | Authors: | Yang, N, Zhang, H, Wang, M, Hao, Q, Sun, H. | | Deposit date: | 2012-09-10 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe.
Sci Rep, 2, 2012
|
|
8WQ3
 
 | | Crystal structure of the C-terminal RRM domain of an RBP | | Descriptor: | CHLORIDE ION, RNA-binding protein 45 | | Authors: | Chen, X, Jiang, M, Yang, Z, Chen, X, Wei, Q, Guo, S, Wang, M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for RNA recognition by the C-terminal RRM domain of human RBM45.
J.Biol.Chem., 300, 2024
|
|
8WQ5
 
 | | Crystal structure of the C-terminal RRM domain of an RBP in complex with ssDNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*GP*CP*AP*G)-3'), GLYCEROL, RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Chen, X, Wei, Q, Guo, S, Jiang, M, Wang, M. | | Deposit date: | 2023-10-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for RNA recognition by the C-terminal RRM domain of human RBM45.
J.Biol.Chem., 300, 2024
|
|
4PW7
 
 | | structure of UHRF2-SRA in complex with a 5mC-containing DNA | | Descriptor: | 5mC-containing DNA1, 5mC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4IMB
 
 | |
4RLM
 
 | | Hen egg-white lysozyme solved from serial crystallography at a synchrotron source, data processed with CrystFEL | | Descriptor: | Lysozyme C | | Authors: | Botha, S, Nass, K, Barends, T, Kabsch, W, Latz, B, Dworkowski, F, Foucar, L, Panepucci, E, Wang, M, Shoeman, R, Schlichting, I, Doak, R.B. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4J12
 
 | | monomeric Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, human Fc fragment | | Authors: | Ishino, T, Wang, M, Mosyak, L, Tam, A, Duan, W, Svenson, K, Joyce, A, O'Hara, D, Lin, L, Somers, W, Kriz, R. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Monomeric Fc Domain Modality by N-Glycosylation for the Half-life Extension of Biotherapeutics.
J.Biol.Chem., 288, 2013
|
|
4IYG
 
 | |
4RLN
 
 | | Hen egg-white lysozyme solved from serial crystallography at a synchrotron source, data processed with nXDS | | Descriptor: | Lysozyme C | | Authors: | Botha, S, Nass, K, Barends, T, Kabsch, W, Latz, B, Dworkowski, F, Foucar, L, Panepucci, E, Wang, M, Shoeman, R, Schlichting, I, Doak, R.B. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
