7YWX
 
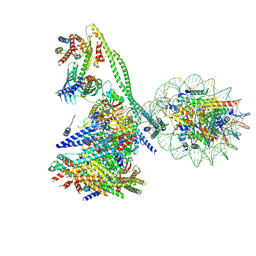 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
3TN4
 
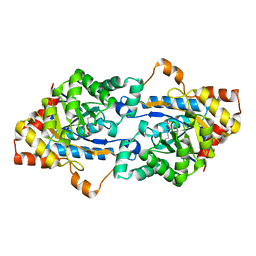 | | Crystal structure of GkaP mutant G209D from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
3TN5
 
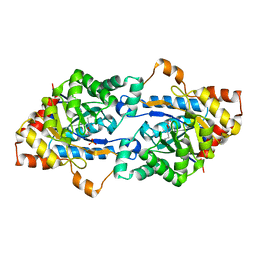 | | Crystal structure of GkaP mutant Y99L from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, PHOSPHATE ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
3TN6
 
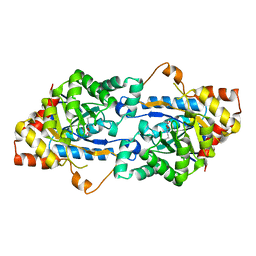 | | Crystal structure of GkaP mutant R230H from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
3TNB
 
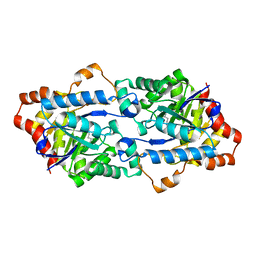 | | Crystal structure of GkaP mutant G209D/R230H from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
8JIN
 
 | |
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVX
 
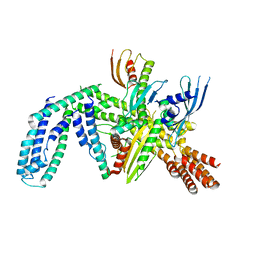 | | Cryo-EM structure of yeast CENP-OPQU+ bound to the CENP-A N-terminus | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CTF19, Inner kinetochore subunit MCM21, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
1RDP
 
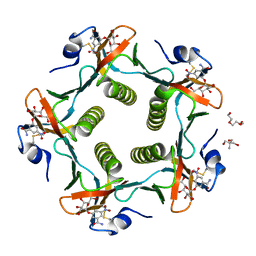 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV3 | | Descriptor: | 1,3-BIS-([[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBU TENYL]-AMINO-ETHYL]-AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RF2
 
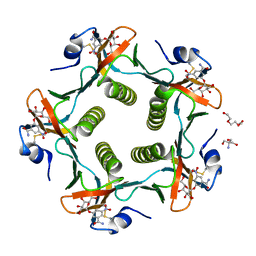 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV4 | | Descriptor: | 1,3-BIS-([3-[3-[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBUTENYL]-AMINO-PROPOXY-ETHOXY-ETHOXY]-PROPYL-]AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-07 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RCV
 
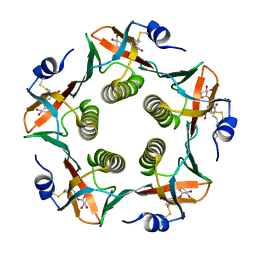 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV1 | | Descriptor: | [3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO] -2-(3-{4-[3-(3-NITRO-5-[GALACTOPYRANOSYLOXY]-BENZOYLAMINO)-PROPYL]-PIPERAZIN-1-YL} -PROPYL-AMINO)-3,4-DIOXO-CYCLOBUTENE, cholera toxin B protein (CTB) | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-04 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
7WZB
 
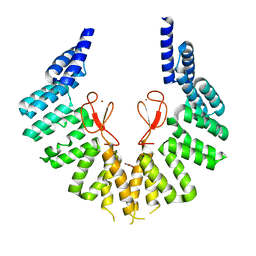 | | lipopolysaccharide assembly protein LapB (open) | | Descriptor: | Lipopolysaccharide assembly protein B, TRIETHYLENE GLYCOL, ZINC ION | | Authors: | Yan, L, Dong, H, Li, H, Liu, X, Deng, Z, Dong, C, Zhang, Z. | | Deposit date: | 2022-02-17 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | lipopolysaccharide assembly protein LapB (open)
To Be Published
|
|
1RYU
 
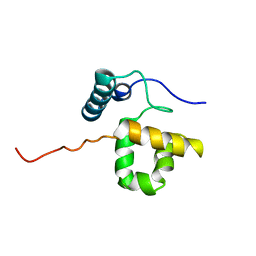 | | Solution Structure of the SWI1 ARID | | Descriptor: | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1 | | Authors: | Kim, S, Zhang, Z, Upchurch, S, Isern, N, Chen, Y. | | Deposit date: | 2003-12-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition.
J.Biol.Chem., 279, 2004
|
|
3TN3
 
 | | Crystal structure of GkaP from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
1RD9
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV2 | | Descriptor: | 1,3-BIS-([3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYL-AMINO]-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1SG0
 
 | | Crystal structure analysis of QR2 in complex with resveratrol | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NRH dehydrogenase [quinone] 2, RESVERATROL, ... | | Authors: | Buryanovskyy, L, Fu, Y, Boyd, M, Ma, Y, Tsieh, T.C, Wu, J.M, Zhang, Z. | | Deposit date: | 2004-02-22 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of quinone reductase 2 in complex with resveratrol
Biochemistry, 43, 2004
|
|
1SCZ
 
 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
3N53
 
 | |
4KQE
 
 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | Descriptor: | GLYCEROL, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
