5CKA
 
 | | Human beta-2 microglobulin double mutant W60G-N83V | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CKG
 
 | | Human beta-2 microglobulin mutant V85E | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CFH
 
 | |
2C7G
 
 | | FprA from Mycobacterium tuberculosis: His57Gln mutant | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-FERREDOXIN REDUCTASE FPRA, ... | | Authors: | Pennati, A, Razeto, A, De Rosa, M, Pandini, V, Vanoni, M.A, Aliverti, A, Mattevi, A, Coda, A, Zanetti, G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the His57-Glu214 Ionic Couple Located in the Active Site of Mycobacterium Tuberculosis Fpra.
Biochemistry, 45, 2006
|
|
7P2B
 
 | |
6QW3
 
 | | Calcium-bound gelsolin domain 2 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Scalone, E, Boni, F, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of gelsolin domain 2 in complex with the physiological calcium ion.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
4BND
 
 | | Structure of an atypical alpha-phosphoglucomutase similar to eukaryotic phosphomannomutases | | Descriptor: | ALPHA-PHOSPHOGLUCOMUTASE, GLYCEROL, SULFATE ION | | Authors: | Nogly, P, Matias, P.M, De Rosa, M, Castro, R, Santos, H, Neves, A.R, Archer, M. | | Deposit date: | 2013-05-14 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-Resolution Structure of an Atypical [Alpha]-Phosphoglucomutase Related to Eukaryotic Phosphomannomutases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6H1F
 
 | | Structure of the nanobody-stabilized gelsolin D187N variant (second domain) | | Descriptor: | Gelsolin, THIOCYANATE ION, gelsolin nanobody, ... | | Authors: | Hassan, A, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
4KA4
 
 | |
5O2Z
 
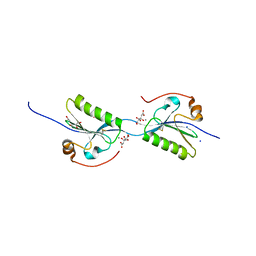 | | Domain swap dimer of the G167R variant of gelsolin second domain | | Descriptor: | ACETATE ION, CALCIUM ION, CITRATE ANION, ... | | Authors: | Boni, F, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gelsolin pathogenic Gly167Arg mutation promotes domain-swap dimerization of the protein.
Hum. Mol. Genet., 27, 2018
|
|
4HOB
 
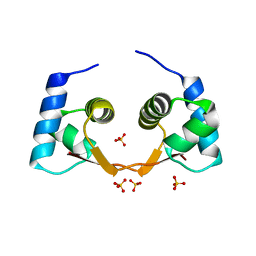 | | The crystal structure of the Zalpha domain from Cyprinid Herpes virus 3 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Tome, A.R, Kus, K, de Rosa, M, Paulo, L.M, Figueiredo, D, Athanasiadis, A. | | Deposit date: | 2012-10-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a poxvirus-like zalpha domain from cyprinid herpesvirus 3
J.Virol., 87, 2013
|
|
4CPW
 
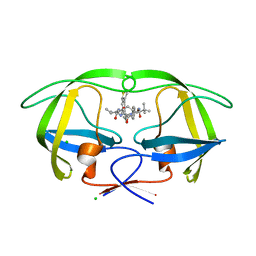 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[[4-[(3S)-3,4-dihydrothiophen-3-yl]phenyl]methyl]-2-[3-[(3Z,8S,11R)-11-oxidanyl-7,10-bis(oxidanylidene)-8-propan-2-yl-6,9-diazabicyclo[11.2.2]heptadeca-1(16),3,13(17),14-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPS
 
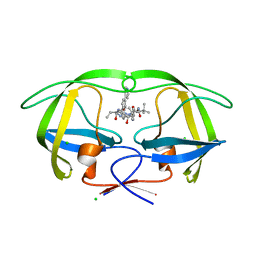 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl [(2S)-1-(2-{3-[(3R,6S,10Z)-3-hydroxy-4,7-dioxo-6-(propan-2-yl)-5,8-diazabicyclo[11.2.2]heptadeca-1(15),10,13,16-tetraen-3-yl]propyl}-2-[4-(pyridin-3-yl)benzyl]hydrazinyl)-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4COE
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl {(2S)-1-[2-(biphenyl-4-ylmethyl)-2-{(4R)-4-hydroxy-5-{[(2S)-3-methyl-1-oxo-1-(prop-2-en-1-ylamino)butan-2-yl]amino}-5-oxo-4-[4-(prop-2-en-1-yl)benzyl]pentyl}hydrazinyl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPQ
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl [(2S)-1-(2-{(4R)-4-hydroxy-5-{[(2S)-3-methyl-1-oxo-1-(prop-2-en-1-ylamino)butan-2-yl]amino}-5-oxo-4-[4-(prop-2-en-1-yl)benzyl]pentyl}-2-[4-(pyridin-4-yl)benzyl]hydrazinyl)-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPT
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl [(2S)-1-(2-{3-[(3R,6S,10Z)-3-hydroxy-4,7-dioxo-6-(propan-2-yl)-5,8-diazabicyclo[11.2.2]heptadeca-1(15),10,13,16-tetraen-3-yl]propyl}-2-[4-(pyridin-3-yl)benzyl]hydrazinyl)-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPX
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl ~{N}-[(2~{S})-3,3-dimethyl-1-[2-[3-[(3~{R},6~{S},10~{Z})-3-oxidanyl-4,7-bis(oxidanylidene)-6-propan-2-yl-5,8-diazabicyclo[11.2.2]heptadeca-1(16),10,13(17),14-tetraen-3-yl]propyl]-2-[(4-thiophen-2-ylphenyl)methyl]hydrazinyl]-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of P1'-functionalized macrocyclic transition-state mimicking HIV-1 protease inhibitors encompassing a tertiary alcohol.
J. Med. Chem., 57, 2014
|
|
4CP7
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl [(2S)-1-(2-{(4R)-4-hydroxy-5-{[(2S)-3-methyl-1-oxo-1-(prop-2-en-1-ylamino)butan-2-yl]amino}-5-oxo-4-[4-(prop-2-en-1-yl)benzyl]pentyl}-2-[4-(pyridin-4-yl)benzyl]hydrazinyl)-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | deRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPR
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl ~{N}-[(2~{S})-3,3-dimethyl-1-[2-[(4~{R})-5-[[(2~{S})-3-methyl-1-oxidanylidene-1-(prop-2-enylamino)butan-2-yl]amino]-4-oxidanyl-5-oxidanylidene-4-[(4-prop-2-enylphenyl)methyl]pentyl]-2-[(4-thiophen-2-ylphenyl)methyl]hydrazinyl]-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPU
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[[4-[(3S)-3,4-dihydrothiophen-3-yl]phenyl]methyl]-2-[3-[(3Z,8S,11R)-11-oxidanyl-7,10-bis(oxidanylidene)-8-propan-2-yl-6,9-diazabicyclo[11.2.2]heptadeca-1(16),3,13(17),14-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
7ZKC
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (apo form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Le Cornu, J.D. | | Deposit date: | 2022-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
7ZLU
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-2-deoxy-2-fluoro-D-glucose | | Descriptor: | 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Ibba, R. | | Deposit date: | 2022-04-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
7ZLL
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with the 5-[(morpholin-4-yl)methyl]quinolin-8-ol inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(morpholin-4-ylmethyl)quinolin-8-ol, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Kantsadi, A.L, Chandran, A.V. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
6EY2
 
 | | Crystal structure of XIAP-BIR3 in complex with a cIAP1-selective SM | | Descriptor: | (3~{S},6~{S},7~{S},9~{a}~{S})-~{N}-[(4-~{tert}-butylphenyl)methyl]-7-(hydroxymethyl)-6-[[(2~{S})-2-(methylamino)butanoyl]amino]-5-oxidanylidene-1,2,3,6,7,8,9,9~{a}-octahydropyrrolo[1,2-a]azepine-3-carboxamide, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Cossu, F, Corti, A, Milani, M, Mastrangelo, E. | | Deposit date: | 2017-11-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based design and molecular profiling of Smac-mimetics selective for cellular IAPs.
FEBS J., 285, 2018
|
|
6EXW
 
 | | Crystal structure of cIAP1-BIR3 in complex with a covalently bound SM | | Descriptor: | (3~{S},6~{S},7~{R},9~{a}~{S})-6-[[(2~{S})-2-(methylamino)propanoyl]amino]-5-oxidanylidene-~{N}-(phenylmethyl)-7-[(propanoylamino)methyl]-3,6,7,8,9,9~{a}-hexahydropyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Corti, A, Cossu, F, Milani, M, Mastrangelo, E. | | Deposit date: | 2017-11-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design and molecular profiling of Smac-mimetics selective for cellular IAPs.
FEBS J., 285, 2018
|
|
