5C54
 
 | |
5U5M
 
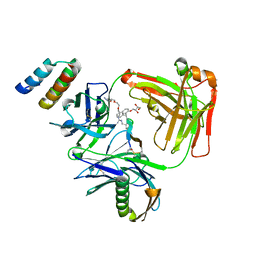 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-MEDITOPE | | Descriptor: | AZIDO-PEG4-MEDITOPE, Immunoglobulin G binding protein A, MEMAB TRASTUZUMAB, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-Meditope
To Be Published
|
|
5U6A
 
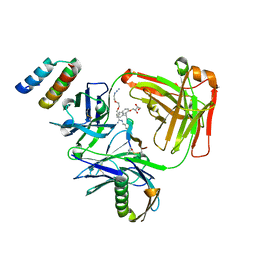 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-PEG3-MEDITOPE | | Descriptor: | Heavy Chain, Immunoglobulin G binding protein A, Light Chain, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-PEG3-Meditope
To Be Published
|
|
2L7U
 
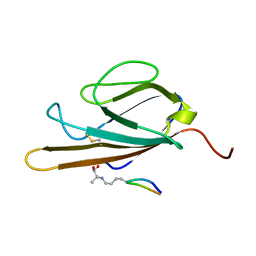 | | Structure of CEL-PEP-RAGE V domain complex | | Descriptor: | Advanced glycosylation end product-specific receptor, Serum albumin peptide | | Authors: | Xue, J, Rai, V, Schmidt, A, Frolov, S, Reverdatto, S, Singer, D, Chabierski, S, Xie, J, Burz, D, Shekhtman, A, Hoffman, R. | | Deposit date: | 2010-12-23 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Advanced glycation end product recognition by the receptor for AGEs.
Structure, 19, 2011
|
|
2MJC
 
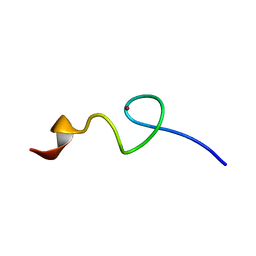 | | Zn-binding domain of eukaryotic translation initiation factor 3, subunit G | | Descriptor: | Eukaryotic translation initiation factor 3 subunit G, ZINC ION | | Authors: | Al-Abdul-Wahid, M, Menade, M, Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Zn-binding domain of eukaryotic translation initiation factor 3, subunit G
To be Published
|
|
8HD6
 
 | | The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8HD7
 
 | | The intermediate pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The intermediate pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8I7N
 
 | | The Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | (2R,3R,4S,5R)-2-(2-azanylpurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7S7P
 
 | |
6N1L
 
 | | The complement inhibitory domain of B. burgdorferi BBK32. | | Descriptor: | Fibronectin-binding protein BBK32 | | Authors: | Garcia, B.L. | | Deposit date: | 2018-11-09 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Structural determination of the complement inhibitory domain of Borrelia burgdorferi BBK32 provides insight into classical pathway complement evasion by lyme disease spirochetes.
PLoS Pathog., 15, 2019
|
|
7CM3
 
 | | Cryo-EM structure of human NALCN in complex with FAM155A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J, Yan, Z, Ke, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human sodium leak channel NALCN in complex with FAM155A.
Nat Commun, 11, 2020
|
|
8TK1
 
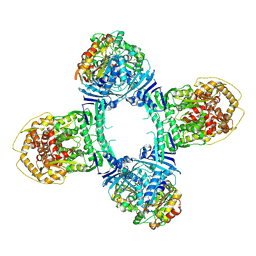 | | Structure of Gabija AB complex 1 | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TK0
 
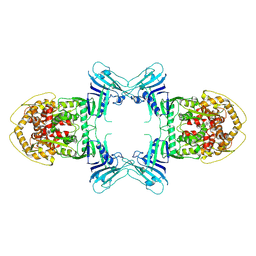 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TJY
 
 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3BWH
 
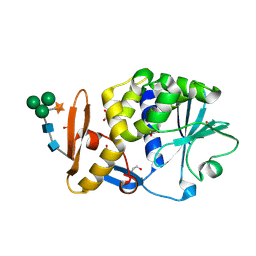 | | Atomic resolution structure of cucurmosin, a novel type 1 RIP from the sarcocarp of Cucurbita moschata | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L. | | Deposit date: | 2008-01-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of cucurmosin, a novel type 1 ribosome-inactivating protein from the sarcocarp of Cucurbita moschata.
J.Struct.Biol., 164, 2008
|
|
5WKO
 
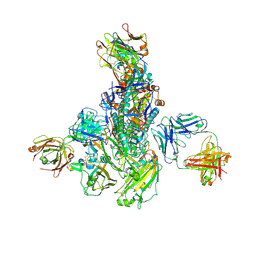 | |
6TYY
 
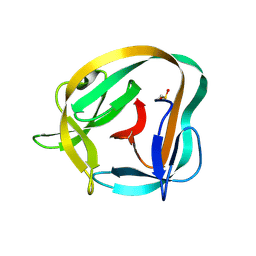 | | Hedgehog autoprocessing mutant D46H | | Descriptor: | Protein hedgehog | | Authors: | Li, H, Li, Z, Wang, C, Callahan, B.P. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | General Base Swap Preserves Activity and Expands Substrate Tolerance in Hedgehog Autoprocessing.
J.Am.Chem.Soc., 141, 2019
|
|
6KQU
 
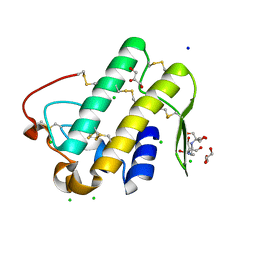 | | Crystal structure of phospholipase A2 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, T, Liu, J. | | Deposit date: | 2019-08-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Residue Asn21 acts as a switch for calcium binding to modulate the enzymatic activity of human phospholipase A2 group IIE.
Biochimie, 176, 2020
|
|
6O9R
 
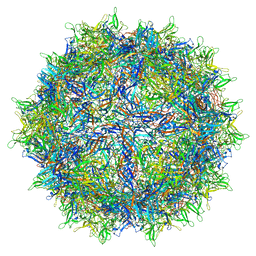 | |
1NIX
 
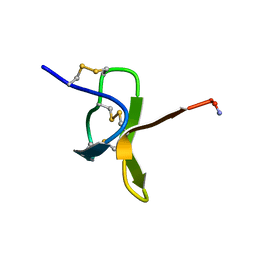 | |
6VQF
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244- 487)-L6-SRC1(678-692)) IN COMPLEX WITH AN INVERSE AGONIST | | Descriptor: | (1R,3S,4R)-4-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]-3-methylcyclohexane-1-carboxylic acid, GLYCEROL, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-02-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of BMS-986251: A Clinically Viable, Potent, and Selective ROR gamma t Inverse Agonist.
Acs Med.Chem.Lett., 11, 2020
|
|
8EEA
 
 | | Structure of E.coli Septu (PtuAB) complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA, PtuB | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EE7
 
 | |
8EE4
 
 | | Structure of PtuA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7MF1
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 47D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 47D1 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Diverse immunoglobulin gene usage and convergent epitope targeting in neutralizing antibody responses to SARS-CoV-2.
Cell Rep, 35, 2021
|
|
