7BPM
 
 | | Solution NMR structure of NF2; de novo designed protein with a novel fold | | Descriptor: | NF2 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQD
 
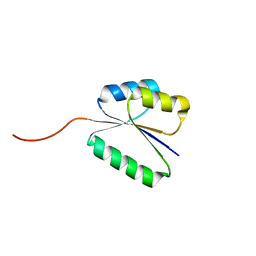 | | Solution NMR structure of NF8 (knot fold); de novo designed protein with a novel fold | | Descriptor: | NF8 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPN
 
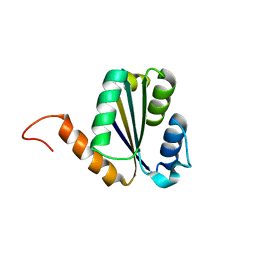 | | Solution NMR structure of NF7; de novo designed protein with a novel fold | | Descriptor: | NF7 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPL
 
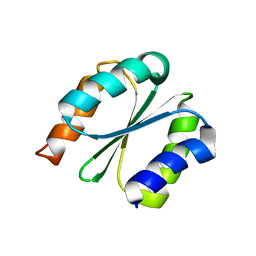 | | Solution NMR structure of NF1; de novo designed protein with a novel fold | | Descriptor: | NF1 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
2D3B
 
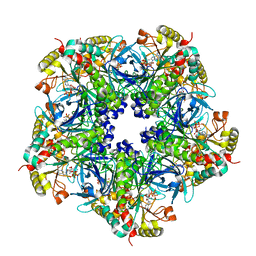 | | Crystal Structure of the Maize Glutamine Synthetase complexed with AMPPNP and Methionine sulfoximine | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2D3A
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with ADP and Methionine sulfoximine Phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2D3C
 
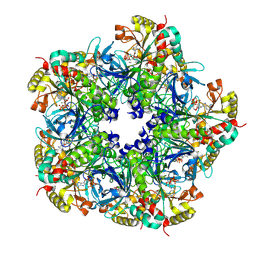 | | Crystal Structure of the Maize Glutamine Synthetase complexed with ADP and Phosphinothricin Phosphate | | Descriptor: | (2S)-2-AMINO-4-[METHYL(PHOSPHONOOXY)PHOSPHORYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
5BOH
 
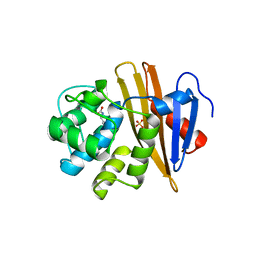 | |
3WBL
 
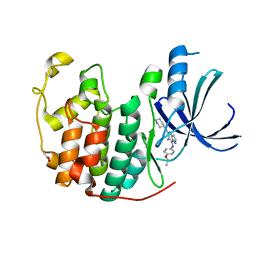 | | Crystal structure of CDK2 in complex with pyrazolopyrimidine inhibitor | | Descriptor: | ACETATE ION, Cyclin-dependent kinase 2, N~7~-(4-ethoxyphenyl)-6-methyl-N~5~-[(3S)-piperidin-3-yl]pyrazolo[1,5-a]pyrimidine-5,7-diamine | | Authors: | Fujino, A, Fukushima, K, Kubota, T, Kosugi, T, Takimoto-Kamimura, M. | | Deposit date: | 2013-05-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase-2 complex with MK2 inhibitor TEI-I01800: insight into the selectivity.
J.SYNCHROTRON RADIAT., 20, 2013
|
|
3ABG
 
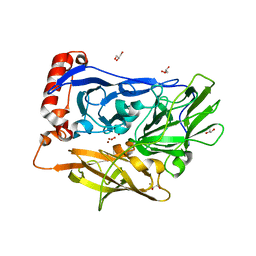 | | X-ray Crystal Analysis of Bilirubin Oxidase from Myrothecium verrucaria at 2.3 angstrom Resolution using a Twin Crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Mizutani, K, Toyoda, M, Sagara, K, Takahashi, N, Sato, A, Kamitaka, Y, Tsujimura, S, Nakanishi, Y, Sugiura, T, Yamaguchi, S, Kano, K, Mikami, B. | | Deposit date: | 2009-12-10 | | Release date: | 2010-08-18 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray analysis of bilirubin oxidase from Myrothecium verrucaria at 2.3 A resolution using a twinned crystal
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4GWG
 
 | |
4GPI
 
 | | Crystal structure of human B type phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Zhou, L, He, C. | | Deposit date: | 2012-08-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0817 Å) | | Cite: | Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation.
Nat Commun, 4, 2013
|
|
4GPZ
 
 | |
4GWK
 
 | | Crystal structure of 6-phosphogluconate dehydrogenase complexed with 3-phosphoglyceric acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PHOSPHOGLYCERIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | He, C, Zhou, L, Zhang, L. | | Deposit date: | 2012-09-03 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth.
Cancer Cell, 22, 2012
|
|
6I7O
 
 | | The structure of a di-ribosome (disome) as a unit for RQC and NGD quality control pathways recognition. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2018-11-16 | | Release date: | 2019-01-16 | | Last modified: | 2019-03-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.
EMBO J., 38, 2019
|
|
4U5X
 
 | | Structure of plant small GTPase OsRac1 complexed with the non-hydrolyzable GTP analog GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ohki, I, Kosami, K, Fujiwara, T, Nakagawa, A, Shimamoto, K, Kojima, C. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of the Plant Small GTPase OsRac1 Reveals Its Mode of Binding to NADPH Oxidase
J.Biol.Chem., 289, 2014
|
|
7QVP
 
 | | Human collided disome (di-ribosome) stalled on XBP1 mRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Denk, T.G, Tesina, P, Beckmann, R. | | Deposit date: | 2022-01-22 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun, 13, 2022
|
|
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6TNU
 
 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6X5Q
 
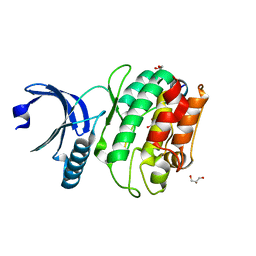 | |
6X5G
 
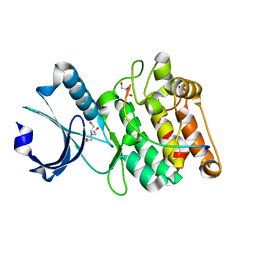 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and LRRC7 inhibitory domain | | Descriptor: | BICINE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
5H6M
 
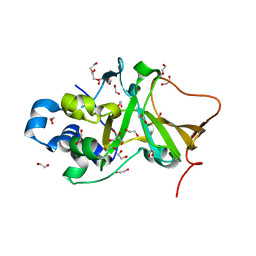 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6J
 
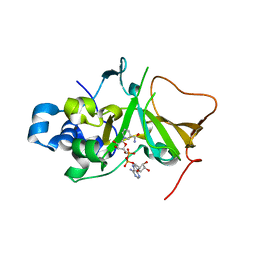 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
