3C3Y
 
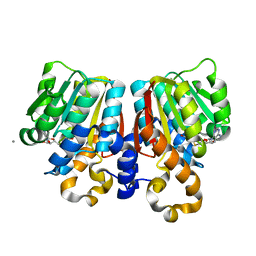 | | Crystal Structure of PFOMT, Phenylpropanoid and Flavonoid O-methyltransferase from M. crystallinum | | Descriptor: | CALCIUM ION, O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kopycki, J.G, Rauh, D, Neumann, P, Stubbs, M.T. | | Deposit date: | 2008-01-29 | | Release date: | 2008-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | Biochemical and Structural Analysis of Substrate Promiscuity in Plant Mg(2+)-Dependent O-Methyltransferases
J.Mol.Biol., 378, 2008
|
|
4WHE
 
 | | Crystal structure of E. coli phage shock protein A (PspA 1-144) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phage shock protein A | | Authors: | Parthier, C, Schoepfel, M, Stubbs, M.T, Osadnik, H, Brueser, T. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PspF-binding domain PspA1-144 and the PspAF complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins.
Mol.Microbiol., 98, 2015
|
|
6GV1
 
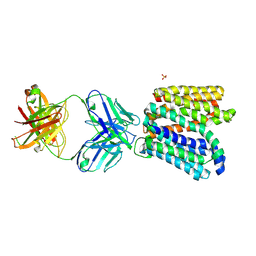 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | Descriptor: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | Authors: | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
8A5T
 
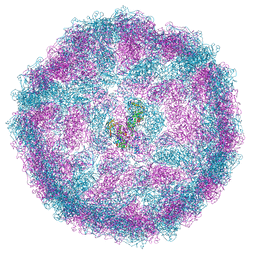 | |
1R5Y
 
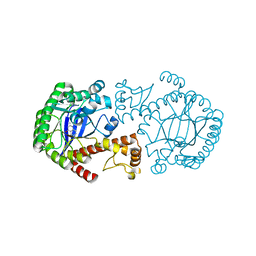 | | Crystal Structure of TGT in complex with 2,6-Diamino-3H-Quinazolin-4-one Crystallized at PH 5.5 | | Descriptor: | 2,6-DIAMINO-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Garcia, G.A, Stubbs, M.T, Klebe, G. | | Deposit date: | 2003-10-13 | | Release date: | 2004-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
2BMA
 
 | | The crystal structure of Plasmodium falciparum glutamate dehydrogenase, a putative target for novel antimalarial drugs | | Descriptor: | GLUTAMATE DEHYDROGENASE (NADP+) | | Authors: | Werner, C, Stubbs, M.T, Krauth-Siege, R.L, Klebe, G. | | Deposit date: | 2005-03-10 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Plasmodium Falciparum Glutamate Dehydrogenase, a Putative Target for Novel Antimalarial Drugs
J.Mol.Biol., 349, 2005
|
|
3FT7
 
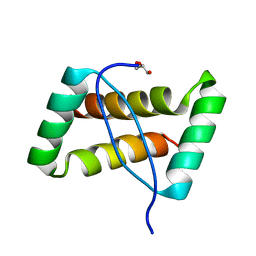 | | Crystal structure of an extremely stable dimeric protein from sulfolobus islandicus | | Descriptor: | GLYCEROL, Uncharacterized protein ORF56 | | Authors: | Neumann, P, Loew, C, Weininger, U, Stubbs, M.T. | | Deposit date: | 2009-01-12 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Stability Analysis of an Extremely Stable Dimeric DNA Binding Protein from Sulfolobus islandicus
Biochemistry, 48, 2009
|
|
1A5I
 
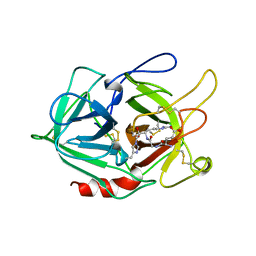 | |
1BTH
 
 | |
5MY4
 
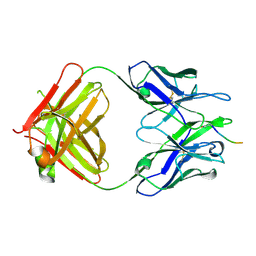 | |
5MYK
 
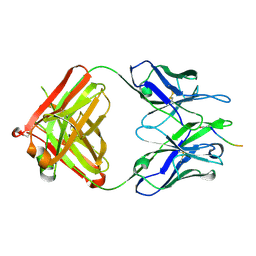 | |
5MYO
 
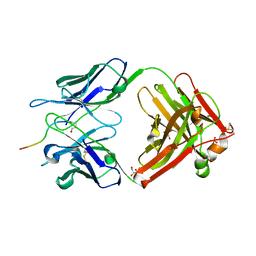 | | Structure of Pyroglutamate-Abeta-specific Fab c#6 in complex with human Abeta-pE3-12-PEGb | | Descriptor: | Amyloid beta A4 protein, Fab c#6 heavy chain, Fab c#6 light chain, ... | | Authors: | Parthier, C, Piechotta, A, Stubbs, M.T. | | Deposit date: | 2017-01-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and functional analyses of pyroglutamate-amyloid-beta-specific antibodies as a basis for Alzheimer immunotherapy.
J. Biol. Chem., 292, 2017
|
|
5MYX
 
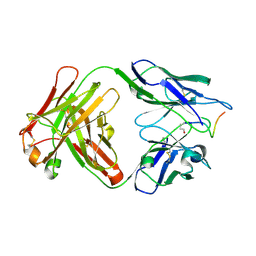 | |
4NIX
 
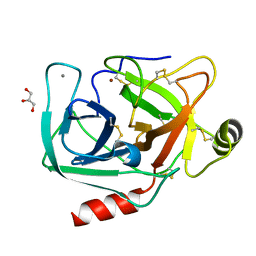 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) orthorhombic form, zinc-bound | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3LN9
 
 | | Crystal structure of the fibril-specific B10 antibody fragment | | Descriptor: | CITRATE ANION, GLYCEROL, Immunoglobulin heavy chain antibody variable domain B10, ... | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Faendrich, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Amyloid Fibril Recognition with the Conformational B10 Antibody Fragment Depends on Electrostatic Interactions.
J.Mol.Biol., 2010
|
|
3LUO
 
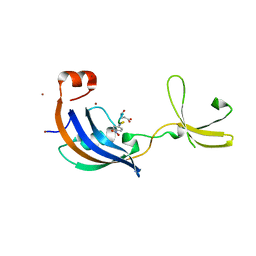 | | Crystal Structure and functional characterization of the thermophilic prolyl isomerase and chaperone SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, Suc-Ala-Leu-Pro-Phe-pNA, ZINC ION | | Authors: | Loew, C, Neumann, P, Weininger, U, Stubbs, M.T, Balbach, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
4NIY
 
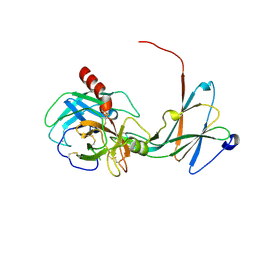 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) complexed to YRH-ecotin (M84Y/M85R/A86H ecotin) | | Descriptor: | CALCIUM ION, Cationic trypsin, Ecotin, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NIV
 
 | |
4NIW
 
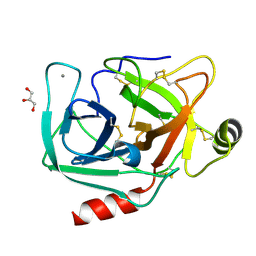 | |
3NOL
 
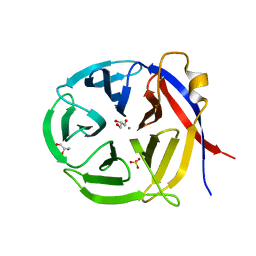 | | Crystal structure of Zymomonas mobilis Glutaminyl Cyclase (trigonal form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
3NOK
 
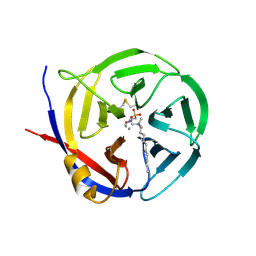 | | Crystal structure of Myxococcus xanthus Glutaminyl Cyclase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
6QRO
 
 | | Crystal structure of Tannerella forsythia glutaminyl cyclase | | Descriptor: | Glutamine cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | Deposit date: | 2019-02-19 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
6QQL
 
 | | Crystal structure of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | Glutamine cyclotransferase, ZINC ION | | Authors: | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
6SHT
 
 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | Descriptor: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | Authors: | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | Deposit date: | 2019-08-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
7YWC
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2022-02-12 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis.
J.Biol.Chem., 298, 2022
|
|
