4LM5
 
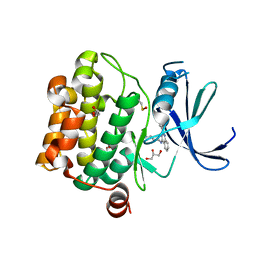 | | Crystal structure of Pim1 in complex with 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol (resulting from displacement of SKF86002) | | Descriptor: | 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-10 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XOG
 
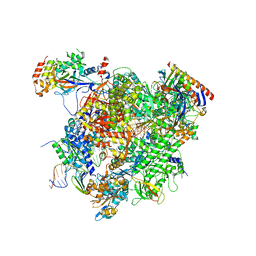 | | RNA Polymerase II elongation complex bound with Spt5 KOW5 and Elf1 | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (30-MER), DNA (39-MER), ... | | Authors: | Ehara, H, Shirouzu, M, Sekine, S. | | Deposit date: | 2017-05-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the complete elongation complex of RNA polymerase II with basal factors
Science, 357, 2017
|
|
4LUD
 
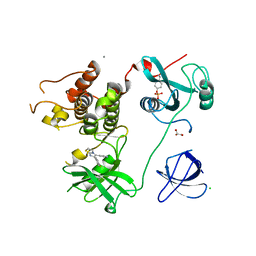 | | Crystal Structure of HCK in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3HJN
 
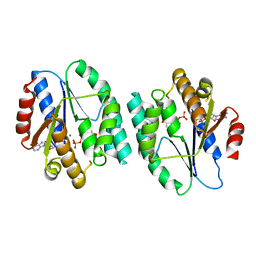 | | Crystal structure of thymidylate kinase in complex with dTDP and ADP from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Yoshikawa, S, Nakagawa, N, Shirouzu, M, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymidylate kinase in complex with dTDP and ADP from Thermotoga maritima
To be Published
|
|
4WR4
 
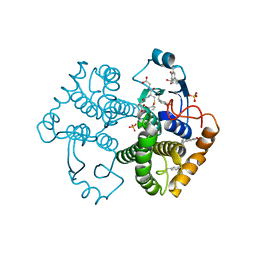 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4WR5
 
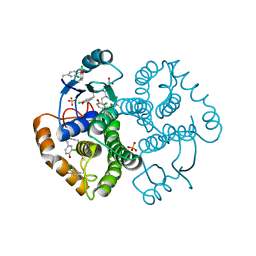 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4LUE
 
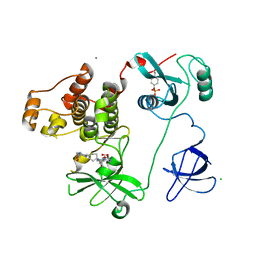 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1RRB
 
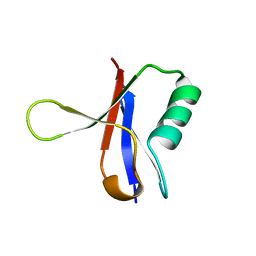 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
1UC9
 
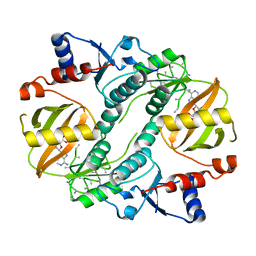 | | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, lysine biosynthesis enzyme | | Authors: | Sakai, H, Vassylyeva, M.N, Matsuura, T, Sekine, S, Nishiyama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a Lysine Biosynthesis Enzyme, LysX, from Thermus thermophilus HB8
J.Mol.Biol., 332, 2003
|
|
8H7X
 
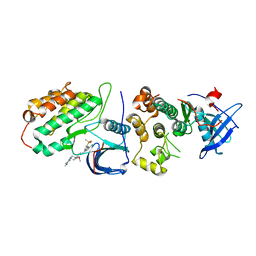 | | Crystal structure of EGFR T790M/C797S mutant in complex with brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | A macrocyclic kinase inhibitor overcomes triple resistant mutations in EGFR-positive lung cancer.
NPJ Precis Oncol, 8, 2024
|
|
9J0P
 
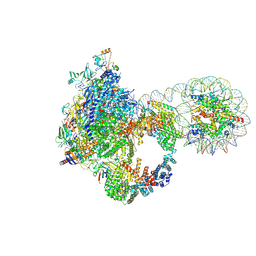 | | Arrested elongation complex of mammalian RNA polymerase II with nucleosome (AEC2-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
9J0N
 
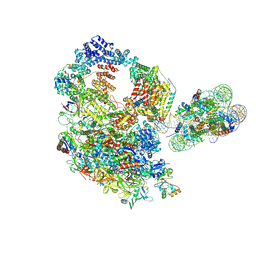 | | Paused elongation complex of mammalian RNA polymerase II with nucleosome (PEC2-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
9J0O
 
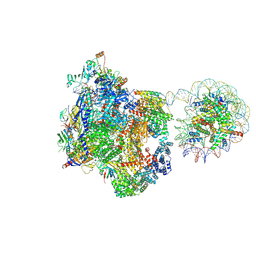 | | Arrested elongation complex of mammalian RNA polymerase II with nucleosome (AEC1-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2025-03-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
3WF8
 
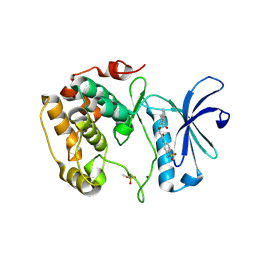 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate | | Descriptor: | 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3WF9
 
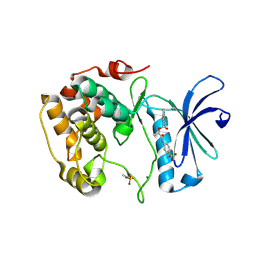 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 1-oxo-1-[(4-sulfamoylphenyl)amino]propan-2-yl-2-methyl-1,2,3,4-tetrahydroacridine-9-carboxylate | | Descriptor: | (2S)-1-oxo-1-[(4-sulfamoylphenyl)amino]propan-2-yl (2S)-2-methyl-1,2,3,4-tetrahydroacridine-9-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3WU4
 
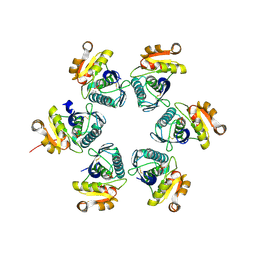 | | Oxidized-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
8H79
 
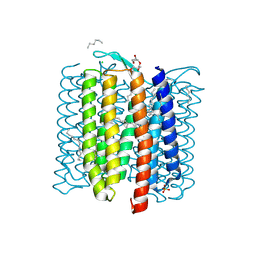 | | The crystal structure of cyanorhodopsin-II (CyR-II) P7104R from Nodosilinea nodulosa PCC 7104 | | Descriptor: | CHLORIDE ION, HEXADECANE, HEXANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2022-10-19 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Cyanorhodopsin-II represents a yellow-absorbing proton-pumping rhodopsin clade within cyanobacteria.
Isme J, 18, 2024
|
|
1NZ8
 
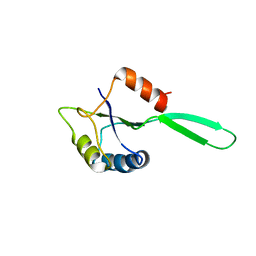 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1NZ9
 
 | | Solution Structure of the N-utilization substance G (NusG) C-terminal (NGC) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
8I5W
 
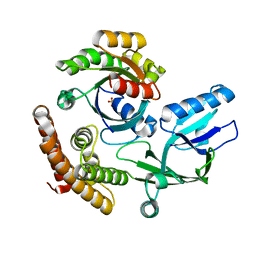 | | Crystal structure of the DHR-2 domain of DOCK10 in complex with Rac1 | | Descriptor: | Dedicator of cytokinesis protein 10, Ras-related C3 botulinum toxin substrate 1, SULFATE ION | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Ihara, K, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.432 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
8I5V
 
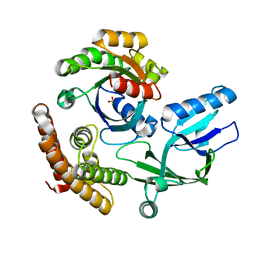 | | DOCK10 mutant L1903Y complexed with Rac1 | | Descriptor: | Dedicator of cytokinesis protein 10, Ras-related C3 botulinum toxin substrate 1, SULFATE ION | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Ihara, K, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
8I5F
 
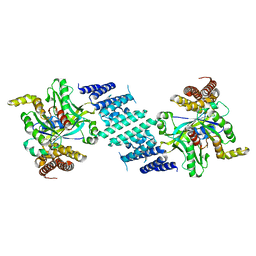 | | Crystal structure of the DHR-2 domain of DOCK10 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 10 | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
5XDN
 
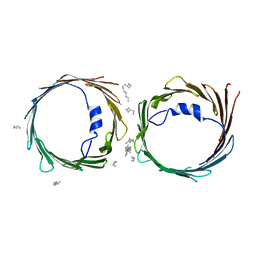 | | Crystal structure of human voltage-dependent anion channel 1 (hVDAC1) in P22121 space group | | Descriptor: | DECANE, DODECANE, HEXANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structural characterization reveals novel oligomeric interactions of human voltage-dependent anion channel 1
Protein Sci., 26, 2017
|
|
