5ZVV
 
 | | Structure of SeMet-phAimR | | Descriptor: | AimR transcriptional regulator, GLYCEROL | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW6
 
 | | Structure of spAimR | | Descriptor: | AimR transcriptional regulator, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW5
 
 | | Structure of SeMet-spAimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-29 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZVW
 
 | | Structure of phAimR-Ligand | | Descriptor: | AimR transcriptional regulator, SER-ALA-ILE-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5GUF
 
 | |
8H18
 
 | |
8H2F
 
 | |
8HI1
 
 | | Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, DNA (26-MER), DNA (31-MER), ... | | Authors: | Chen, Q, Luo, Y. | | Deposit date: | 2022-11-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | DnaQ mediates directional spacer acquisition in the CRISPR-Cas system by a time-dependent mechanism.
Innovation (N Y), 4, 2023
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
4FWJ
 
 | | Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, PHOSPHATE ION, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWE
 
 | | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWF
 
 | | Complex structure of LSD2/AOF1/KDM1b with H3K4 mimic | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.1, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
6OCG
 
 | | Crystal structure of VASH1-SVBP complex bound with EpoY | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[(3R)-4-ethoxy-3-hydroxy-4-oxobutanoyl]-L-tyrosine, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8J1V
 
 | |
8J1T
 
 | |
6J1O
 
 | |
6J46
 
 | | LepI-SAH complex structure | | Descriptor: | O-methyltransferase lepI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
6J24
 
 | | Crystal structure of a SAM-dependent methyltransferase LepI in complex with its substrate | | Descriptor: | (3~{S},4'~{R},4'~{a}~{S},6'~{R},8'~{a}~{S})-4',6'-dimethyl-5-phenyl-spiro[1~{H}-pyridine-3,5'-2,3,4,4~{a},6,8~{a}-hexahydro-1~{H}-naphthalene]-2,4-dione, O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2018-12-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5F1K
 
 | | human CD38 in complex with nanobody MU1053 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU1053 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5F1O
 
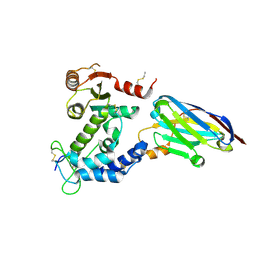 | | human CD38 in complex with nanobody MU551 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU551 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
4DRA
 
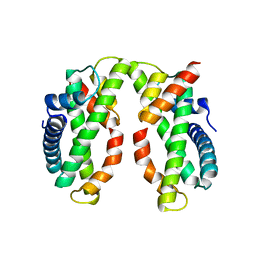 | | Crystal structure of MHF complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | The structure of the FANCM-MHF complex reveals physical features for functional assembly
Nat Commun, 3, 2012
|
|
4DT1
 
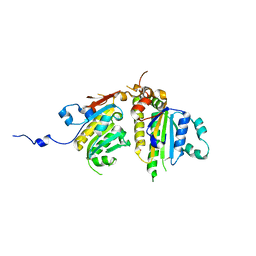 | | Crystal structure of the Psy3-Csm2 complex | | Descriptor: | Chromosome segregation in meiosis protein 2, ETHANOL, Platinum sensitivity protein 3 | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-20 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage
J.Biol.Chem., 287, 2012
|
|
4DRB
 
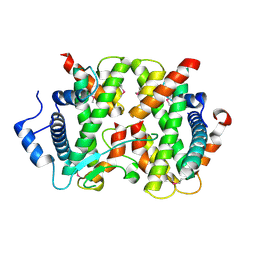 | | The crystal structure of FANCM bound MHF complex | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | The structure of the FANCM-MHF complex reveals physical features for functional assembly
Nat Commun, 3, 2012
|
|
