4K96
 
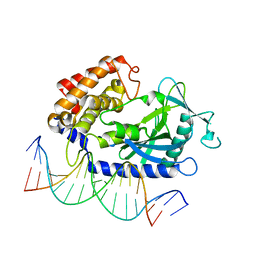 | | Structure of Binary Complex of cGAS with Bound dsDNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA-F, DNA-R, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K98
 
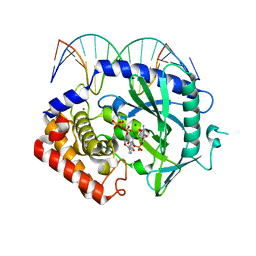 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, DNA-F, DNA-R, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
2RI5
 
 | |
4KF6
 
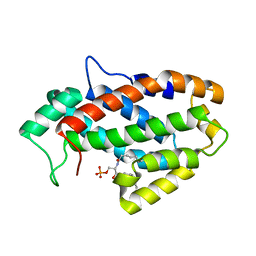 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 8:0 Ceramide-1-Phosphate (8:0-C1P) | | Descriptor: | (2S,3R,4E)-3-hydroxy-2-(octanoylamino)octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K85
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 Ceramide-1-Phosphate (12:0-C1P) | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K8N
 
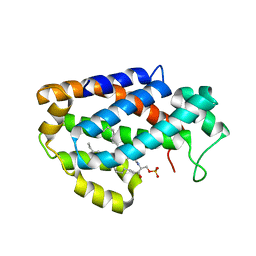 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 18:1 Ceramide-1-Phosphate (18:1-C1P) | | Descriptor: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
2RHU
 
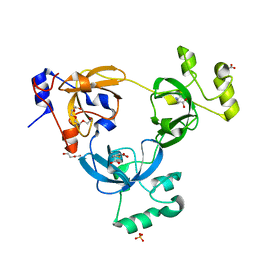 | |
2RI7
 
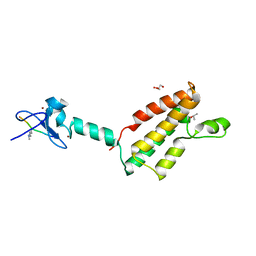 | |
2RI2
 
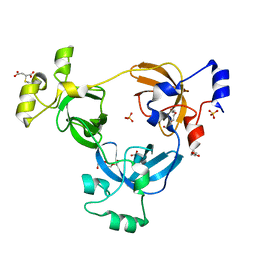 | |
4K99
 
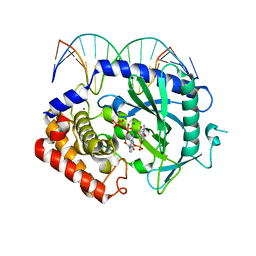 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppdG(2 ,5 )pdG | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 3'-deoxy-guanosine 5'-monophosphate, Cyclic GMP-AMP synthase, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
2RHI
 
 | |
2RHY
 
 | |
9MUE
 
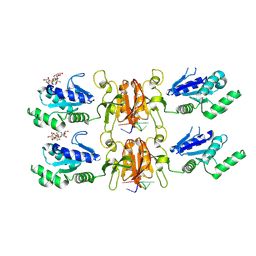 | | Cryo-EM structure of CRISPR-associated cA4 bound Cat1 Pentagonal filament assembly in the presence of NAD (ADPR modelled) | | Descriptor: | Cat1 (CRISPR associated TIR 1) pentagonal filament assembly, RNA (5'-R(P*AP*AP*AP*A)-3'), [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL[HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2025-01-13 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cat1 forms filament networks to degrade NAD + during the type III CRISPR-Cas antiviral response.
Science, 388, 2025
|
|
9MUO
 
 | | Cryo-EM structure of CRISPR-associated cA4 bound Cat1 Pentagonal filament assembly in the presence of NAD analog BAD | | Descriptor: | Cat1 (CRISPR-associated TIR 1), RNA (5'-R(P*AP*AP*AP*A)-3'), [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylphenyl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2025-01-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cat1 forms filament networks to degrade NAD + during the type III CRISPR-Cas antiviral response.
Science, 388, 2025
|
|
9MW9
 
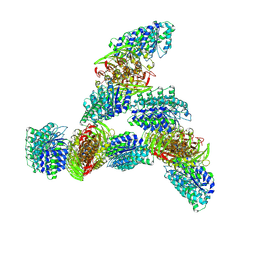 | |
8URQ
 
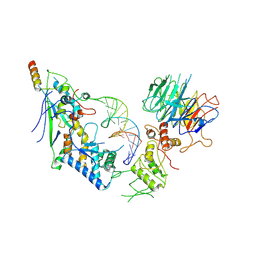 | | Spo11 core complex with gapped DNA | | Descriptor: | Antiviral protein SKI8, MAGNESIUM ION, Meiosis-specific protein SPO11, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8URU
 
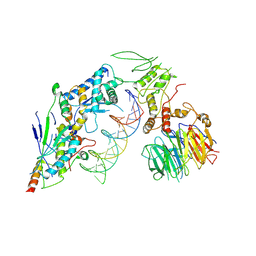 | | Spo11 core complex with hairpin DNA | | Descriptor: | Antiviral protein SKI8, Hairpin DNA, MAGNESIUM ION, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5KDI
 
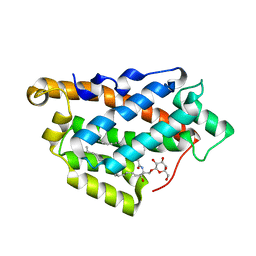 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
7DMQ
 
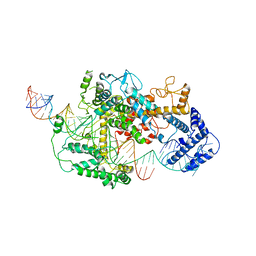 | | Cryo-EM structure of LshCas13a-crRNA-anti-tag RNA complex | | Descriptor: | Anti-tag target RNA, CRISPR RNA, CRISPR/Cas system Cas13a | | Authors: | Wang, B, Zhang, T, Ding, J, Patel, D.J, Yang, H. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.
Mol.Cell, 81, 2021
|
|
7WRN
 
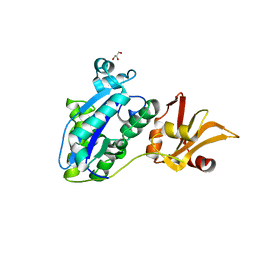 | |
7VKJ
 
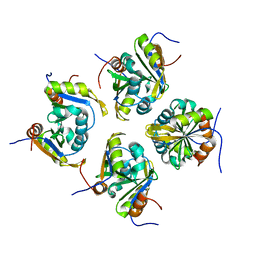 | | Structure of ESRP1 qRRM3 domain | | Descriptor: | Epithelial splicing regulatory protein 1 | | Authors: | Wu, B.X, Patel, D.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of ESRP1 qRRM3 domain
To Be Published
|
|
3QLC
 
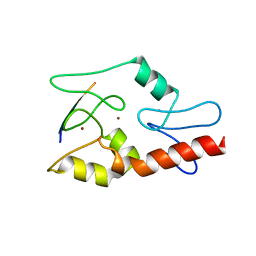 | |
3QLN
 
 | | Crystal structure of ATRX ADD domain in free state | | Descriptor: | Transcriptional regulator ATRX, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
4NJ5
 
 | | Crystal structure of SUVH9 | | Descriptor: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
8T64
 
 | | Apo Cam1(42-206) | | Descriptor: | Cam1 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
