8QKS
 
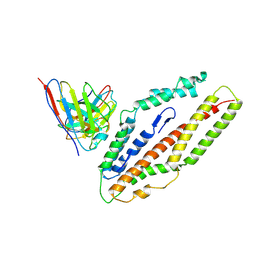 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to R5.034 | | Descriptor: | Immunoglobulin lambda variable 1-36, R5034HV, Reticulocyte-binding protein-like protein 5 | | Authors: | Wright, N.D, Barrett, J.R, Bradshaw, W.J, Paterson, N.G, MacLean, E.M, Ferreira, L, McHugh, K, Von Delft, F, Koekemoer, L, Draper, S.J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.994 Å) | | Cite: | Analysis of the diverse antigenic landscape of the malaria protein RH5 identifies a potent vaccine-induced human public antibody clonotype.
Cell, 187, 2024
|
|
8QKR
 
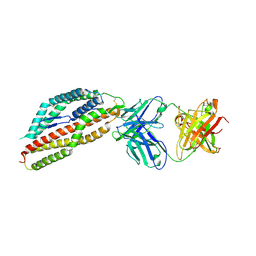 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to R5.251 | | Descriptor: | R5251VHCH, R5251VLCL, Reticulocyte-binding protein-like protein 5 | | Authors: | Wright, N.D, Barrett, J.R, Bradshaw, W.J, Paterson, N.G, MacLean, E.M, Ferreira, L, McHugh, K, Koekemoer, L, Draper, S.J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.234 Å) | | Cite: | Analysis of the diverse antigenic landscape of the malaria protein RH5 identifies a potent vaccine-induced human public antibody clonotype.
Cell, 187, 2024
|
|
7AIO
 
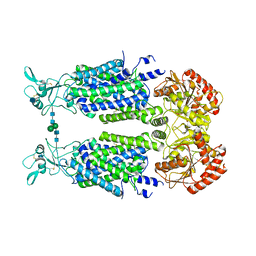 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Subclass) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
7AIN
 
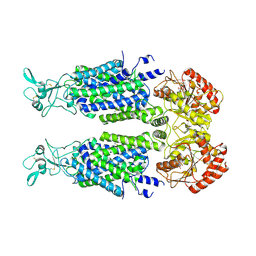 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Reference Map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
7AIR
 
 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Subclass 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Liko, I, Tehan, B.G, Almeida, F.G, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
7AIQ
 
 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Subclass 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Solute carrier family 12 member 4, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Liko, I, Tehan, B.G, Almeida, F.G, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
7AIP
 
 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Reference Map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Liko, I, Tehan, B.G, Almeida, F.G, Elkins, J, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
8OUO
 
 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | Descriptor: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | Deposit date: | 2023-04-24 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 32, 2024
|
|
8P2W
 
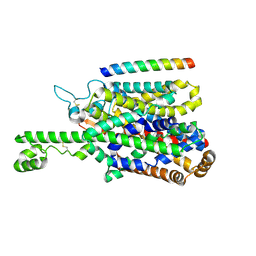 | | Structure of human SIT1 (focussed map / refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Sodium- and chloride-dependent transporter XTRP3 | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2Z
 
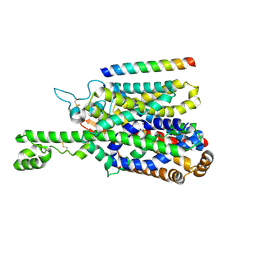 | | Structure of human SIT1 bound to L-pipecolate (focussed map / refinement) | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P30
 
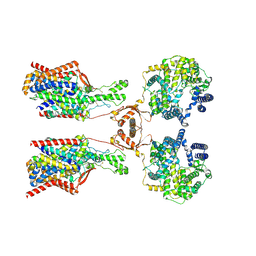 | | Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2Y
 
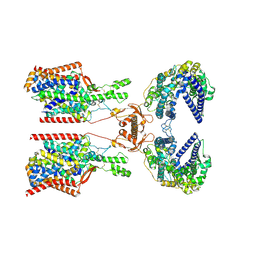 | | Structure of human SIT1:ACE2 complex (closed PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P31
 
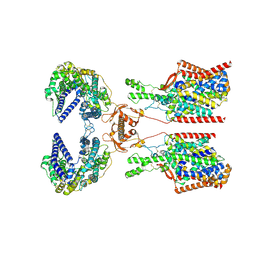 | | Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2X
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8BLO
 
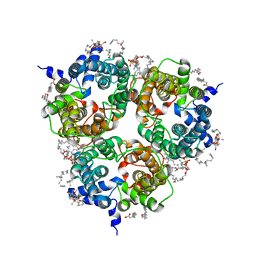 | | Human Urea Transporter UT-A (N-Terminal Domain Model) | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Urea transporter 2, di-heneicosanoyl phosphatidyl choline | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Scacioc, A, Wang, D, McKinley, G, Fernandez-Cid, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Burgess-Brown, N.A, van Putte, W, Duerr, K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
8BLP
 
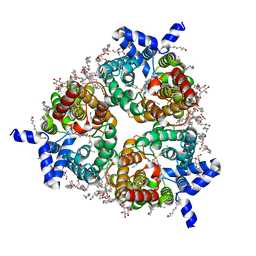 | | Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14 | | Descriptor: | 10-(4-ethylphenyl)sulfonyl-~{N}-(thiophen-2-ylmethyl)-5-thia-1,8,11,12-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,7,9,11-pentaen-7-amine, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Chi, G, Dietz, L, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, Scacioc, A, McKinley, G, Arrowsmith, C.H, Edwards, A, Bountra, C, Fernandez-Cid, A, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
7GB6
 
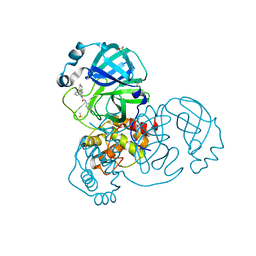 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JOR-UNI-2fc98d0b-12 (Mpro-x10236) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-N-(2-cyclohexylethyl)-N'-(pyridin-3-yl)urea | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBN
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with RAL-MED-2de63afb-14 (Mpro-x10387) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-fluoro-5-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GC2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-93268d01-13 (Mpro-x10484) | | Descriptor: | 3-methyl-N-(4-methylpyridin-3-yl)-3-phenylbutanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB8
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-14 (Mpro-x10247) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-[3-(trifluoromethyl)phenyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBK
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-95b75b4d-2 (Mpro-x10359) | | Descriptor: | 2-(3-hydroxyphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCN
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-41 (Mpro-x10679) | | Descriptor: | 2-(6-chloro-1H-indol-1-yl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-c59291d4-3 (Mpro-x10870) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[4-[2-(benzotriazol-1-yl)ethanoyl-(thiophen-3-ylmethyl)amino]phenyl]cyclopropanecarboxamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBY
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-590ac91e-5 (Mpro-x10473) | | Descriptor: | (2R)-3-cyclopropyl-2-methyl-N-(4-methylpyridin-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCB
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-13 (Mpro-x10565) | | Descriptor: | 2-(3-iodophenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
