8JDF
 
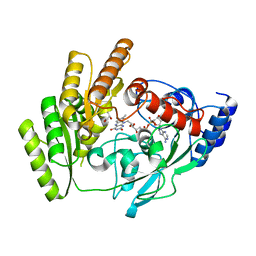 | | Crystal structure of H405A mLDHD in complex with D-lactate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDX
 
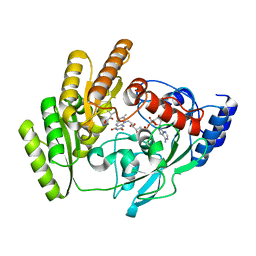 | | Crystal structure of mLDHD in complex with 2-ketoisovaleric acid | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDE
 
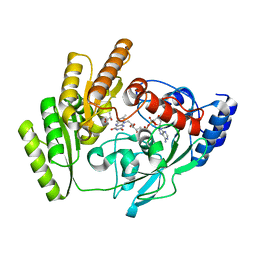 | | Crystal structure of mLDHD in complex with D-lactate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDG
 
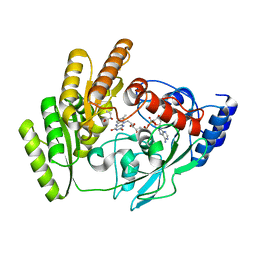 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxybutanoic acid | | Descriptor: | (2R)-2-oxidanylbutanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDR
 
 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxy-3-methyl-valeric acid | | Descriptor: | (2R,3S)-3-methyl-2-oxidanyl-pentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDU
 
 | | Crystal structure of mLDHD in complex with 2-ketovaleric acid | | Descriptor: | 2-oxopentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDY
 
 | | Crystal structure of mLDHD in complex with 2-ketoisocaproic acid | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDO
 
 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyhexanoic acid | | Descriptor: | (2R)-2-hydroxyhexanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDP
 
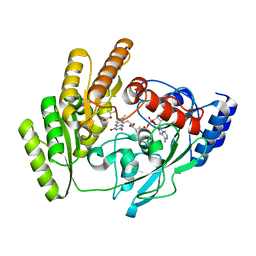 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyisovaleric acid | | Descriptor: | DEAMINOHYDROXYVALINE, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
5FEK
 
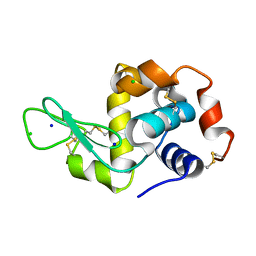 | |
8IQA
 
 | |
5FDJ
 
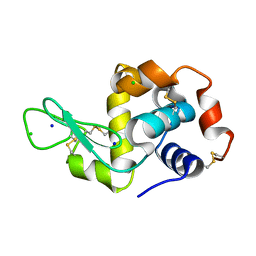 | |
5FEL
 
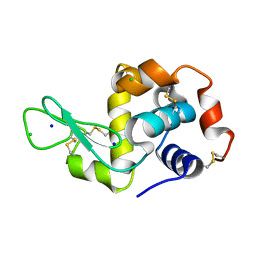 | |
8IL8
 
 | |
8IX6
 
 | |
4DQM
 
 | | Revealing a marine natural product as a novel agonist for retinoic acid receptors with a unique binding mode and antitumor activity | | Descriptor: | (5S)-4-[(3E,7E)-4,8-dimethyl-10-(2,6,6-trimethylcyclohex-1-en-1-yl)deca-3,7-dien-1-yl]-5-hydroxyfuran-2(5H)-one, Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | Authors: | Wang, S, Wang, Z, Lin, S, Zheng, W, Wang, R, Jin, S, Chen, J, Jin, L, Li, Y. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Revealing a natural marine product as a novel agonist for retinoic acid receptors with a unique binding mode and inhibitory effects on cancer cells.
Biochem.J., 446, 2012
|
|
9JE0
 
 | | Human URAT1 bound to benzbromarone | | Descriptor: | Solute carrier family 22 member 12, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JE1
 
 | | Human URAT1 bound to dotinurad | | Descriptor: | Solute carrier family 22 member 12, dotinurad | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JDY
 
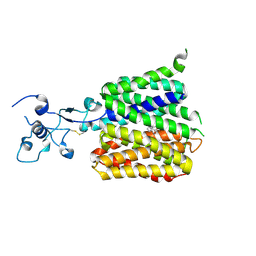 | | Human URAT1 bound with verinurad | | Descriptor: | Solute carrier family 22 member 12, verinurad | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JDV
 
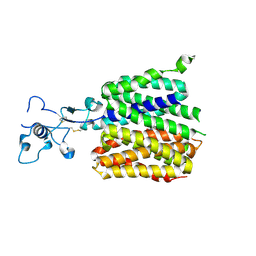 | | Human URAT1 bound with Uric acid | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
1MRJ
 
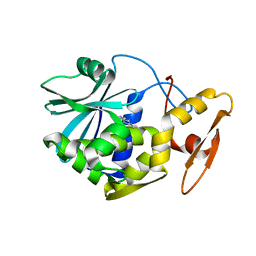 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRH
 
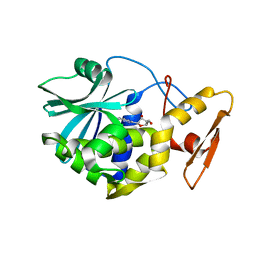 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRK
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRG
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRI
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
