1FAJ
 
 | | INORGANIC PYROPHOSPHATASE | | Descriptor: | SOLUBLE INORGANIC PYROPHOSPHATASE | | Authors: | Kankare, J.A, Salminen, T, Goldman, A. | | Deposit date: | 1996-01-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Escherichia coli inorganic pyrophosphatase at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1YPP
 
 | | ACID ANHYDRIDE HYDROLASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Harutyunyan, E.H, Kuranova, I.P, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 1996-05-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate.
Eur.J.Biochem., 239, 1996
|
|
3ZUV
 
 | |
3ZU7
 
 | |
6G70
 
 | | Structure of murine Prpf39 | | Descriptor: | Pre-mRNA-processing factor 39 | | Authors: | De Bortoli, F.D, Loll, B, Wahl, M, Heyd, F. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Increased versatility despite reduced molecular complexity: evolution, structure and function of metazoan splicing factor PRPF39.
Nucleic Acids Res., 47, 2019
|
|
4TSE
 
 | |
4C59
 
 | | Structure of GAK kinase in complex with nanobody (NbGAK_4) | | Descriptor: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, Cyclin-G-associated kinase, NANOBODY | | Authors: | Chaikuad, A, Keates, T, Allerston, C.K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Muller-Knapp, S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies.
Biochem. J., 459, 2014
|
|
4C58
 
 | | Structure of GAK kinase in complex with nanobody (NbGAK_4) | | Descriptor: | 1,2-ETHANEDIOL, 9-HYDROXY-4-PHENYLPYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Keates, T, Allerston, C.K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Muller-Knapp, S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies.
Biochem. J., 459, 2014
|
|
4C57
 
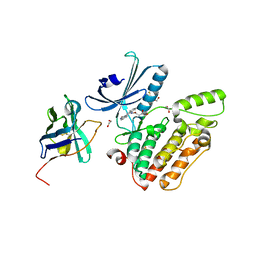 | | Structure of GAK kinase in complex with a nanobody | | Descriptor: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, 1,2-ETHANEDIOL, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Keates, T, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Muller-Knapp, S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Cyclin G-Associated Kinase (Gak) Trapped in Different Conformations Using Nanobodies.
Biochem.J., 459, 2014
|
|
8SOS
 
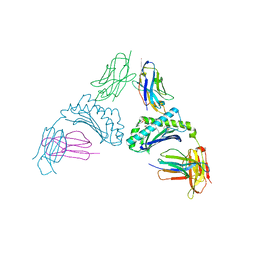 | | Human CD1d presenting sphingomyelin C24:1 in complex with VHH nanobody 1D17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, Beta-2-microglobulin, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2023-04-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Enhanced CD1d phosphatidylserine presentation using a single-domain antibody promotes immunomodulatory CD1d-TIM-3 interactions.
J Immunother Cancer, 11, 2023
|
|
6BDZ
 
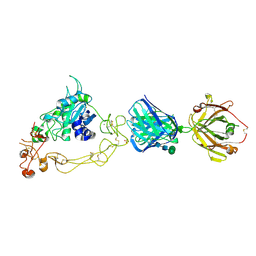 | | ADAM10 Extracellular Domain Bound by the 11G2 Fab | | Descriptor: | 11G2 Fab Heavy Chain, 11G2 Fab Light Chain, CALCIUM ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
6BE6
 
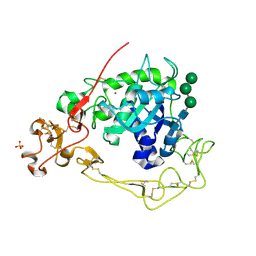 | | ADAM10 Extracellular Domain | | Descriptor: | CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 10, SULFATE ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
5JEA
 
 | | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Kowalinski, E, Ebert, J, Stegmann, E, Conti, E. | | Deposit date: | 2016-04-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex.
Mol.Cell, 63, 2016
|
|
5JNB
 
 | | structure of GLD-2/RNP-8 complex | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Poly(A) RNA polymerase gld-2, ... | | Authors: | Nakel, K, Bonneau, F, Basquin, C, Eckmann, C.R, Conti, E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Structural basis for the antagonistic roles of RNP-8 and GLD-3 in GLD-2 poly(A)-polymerase activity.
Rna, 22, 2016
|
|
6G0I
 
 | | Active Fe-PP1 | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2018-03-18 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of stably incorporated iron on protein phosphatase-1 structure and activity.
FEBS Lett., 592, 2018
|
|
6G0J
 
 | | Inactive Fe-PP1 | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2018-03-18 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of stably incorporated iron on protein phosphatase-1 structure and activity.
FEBS Lett., 592, 2018
|
|
1LDT
 
 | | COMPLEX OF LEECH-DERIVED TRYPTASE INHIBITOR WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPTASE INHIBITOR | | Authors: | Stubbs, M.T. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of recombinant leech-derived tryptase inhibitor in complex with trypsin. Implications for the structure of human mast cell tryptase and its inhibition.
J.Biol.Chem., 272, 1997
|
|
5T9J
 
 | | Crystal Structure of human GEN1 in complex with Holliday junction DNA in the upper interface | | Descriptor: | DNA (5'-D(*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DCP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DAP*DTP*DTP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DAP*DGP*DGP*DGP*DAP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DAP*DGP*DCP*DCP*DTP*DAP*DGP*DCP*DGP*DTP*DCP*DCP*DGP*DGP*DAP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Lee, S.-H, Biertumpfel, C. | | Deposit date: | 2016-09-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.00012732 Å) | | Cite: | Human Holliday junction resolvase GEN1 uses a chromodomain for efficient DNA recognition and cleavage.
Elife, 4, 2015
|
|
1H6F
 
 | | Human TBX3, a transcription factor responsible for ulnar-mammary syndrome, bound to a palindromic DNA site | | Descriptor: | 5'-D(*TP*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP* AP*GP*GP*TP*GP*TP*GP*AP*AP*AP*T)-3', MAGNESIUM ION, T-BOX TRANSCRIPTION FACTOR TBX3 | | Authors: | Coll, M, Muller, C.W. | | Deposit date: | 2001-06-13 | | Release date: | 2002-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the DNA-Bound T-Box Domain of Human Tbx3, a Transcription Factor Responsible for Ulnar- Mammary Syndrome
Structure, 10, 2002
|
|
5UX6
 
 | | Structure of Human POFUT1 in its apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL | | Authors: | Xu, X, McMillan, B, Blacklow, S.C. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations.
Glycobiology, 27, 2017
|
|
5UXH
 
 | | Structure of Human POFUT1 in complex with GDP-fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE | | Authors: | Xu, X, McMillan, B, Blacklow, S.C. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations.
Glycobiology, 27, 2017
|
|
6GN5
 
 | | CRYSTAL STRUCTURE OF HUMAN GRAMD1C START DOMAIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GRAM domain-containing protein 1C | | Authors: | Friese, A, Vetter, I.R. | | Deposit date: | 2018-05-30 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis.
Nat.Chem.Biol., 15, 2019
|
|
2OO9
 
 | |
6ZK6
 
 | | Protein Phosphatase 1 (PP1) T320E mutant | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards Dissecting the Mechanism of Protein Phosphatase-1 Inhibition by Its C-Terminal Phosphorylation.
Chembiochem, 22, 2021
|
|
