5C3Q
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and thymine (T) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, NICKEL (II) ION, ... | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3O
 
 | | Crystal structure of the C-terminal truncated Neurospora crassa T7H (NcT7HdeltaC) in apo form | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Thymine dioxygenase | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3R
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-hydroxymethyluracil (5hmU) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 5-HYDROXYMETHYL URACIL, ... | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3S
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-formyluracil (5fU) | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carbaldehyde, 2-OXOGLUTARIC ACID, ... | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3P
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, NICKEL (II) ION, ... | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C1Q
 
 | | Serine/threonine-protein kinase pim-1 | | Descriptor: | 3-methoxy[1]benzothieno[2,3-c]quinolin-6(5H)-one, Serine/threonine-protein kinase pim-1 | | Authors: | Li, W, Wan, X, Huang, N. | | Deposit date: | 2015-06-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Serine/threonine-protein kinase pim-1
To Be Published
|
|
3IZP
 
 | |
6OLI
 
 | |
6OLF
 
 | |
6OM0
 
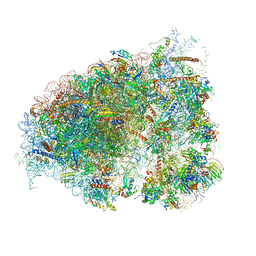 | |
4D3Z
 
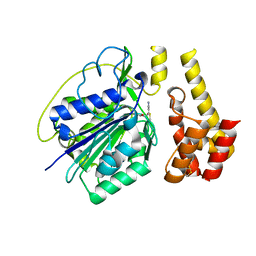 | |
4D3X
 
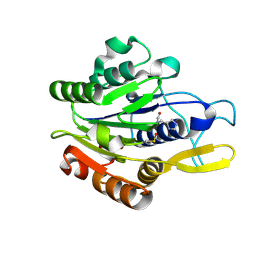 | |
4D3Y
 
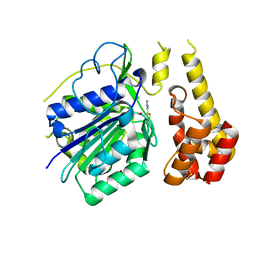 | |
3TJT
 
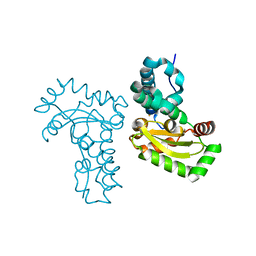 | | Crystal Structure Analysis of the superoxide dismutase from Clostridium difficile | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Lei, C, Ying, T.L, Wang, H.F, Tan, X.S. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of the superoxide dismutase from Clostridium difficile
To be Published
|
|
5Y6R
 
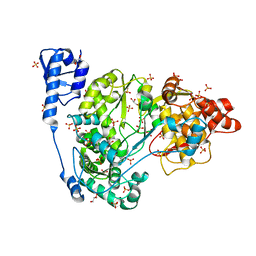 | | Crystal structure of CSFV NS5B | | Descriptor: | GLYCEROL, Genome polyprotein, SULFATE ION | | Authors: | Li, W, Wu, B. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Classical Swine Fever Virus NS5B Reveals a Novel N-Terminal Domain
J. Virol., 92, 2018
|
|
4JZG
 
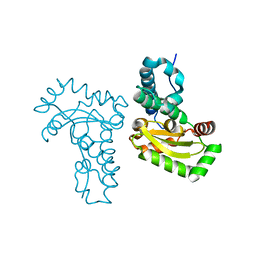 | | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Li, W, Wang, C.L, Zhao, Y, Wang, H.F, Tan, S.X. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile
To be Published
|
|
4JZ2
 
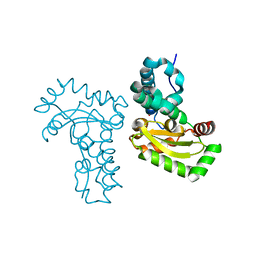 | | Crystal structure of Co ion substituted SOD2 from Clostridium difficile | | Descriptor: | COBALT (II) ION, Superoxide dismutase | | Authors: | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Co ion substituted SOD2 from Clostridium difficile
To be Published
|
|
4JYY
 
 | | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex | | Descriptor: | AZIDE ION, FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex
To be Published
|
|
7E2G
 
 | | Cryo-EM structure of hDisp1NNN-3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1,Protein dispatched homolog 1 | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
7E2I
 
 | | Cryo-EM structure of hDisp1NNN-ShhN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1, ... | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
7E2H
 
 | | Cryo-EM structure of hDisp1NNN-3C-Cleavage | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
6JCI
 
 | |
7CRP
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
