5MN8
 
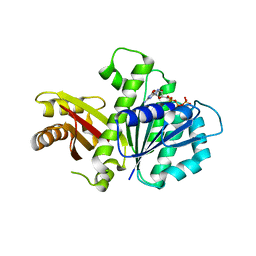 | | S. aureus FtsZ 12-316 F138A GTP Closed form (5FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
3ZIV
 
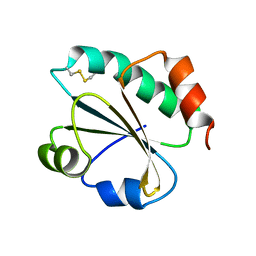 | |
5MN5
 
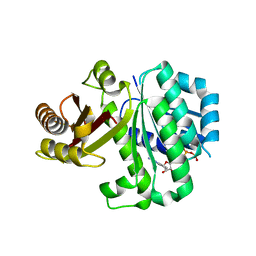 | | S. aureus FtsZ 12-316 T66W GTP Closed form (2TCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN6
 
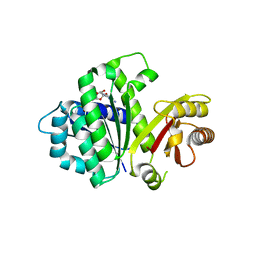 | | S. aureus FtsZ 12-316 F138A GDP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
2IJQ
 
 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
1JR5
 
 | | Solution Structure of the Anti-Sigma Factor AsiA Homodimer | | Descriptor: | 10 KDA Anti-Sigma Factor | | Authors: | Urbauer, J.L, Simeonov, M.F, Bieber Urbauer, R.J, Adelman, K, Gilmore, J.M, Brody, E.N. | | Deposit date: | 2001-08-10 | | Release date: | 2002-02-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the anti-sigma factor AsiA: implications for novel functions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6SYJ
 
 | |
3FK3
 
 | | Structure of the Yeats Domain, Yaf9 | | Descriptor: | Protein AF-9 homolog | | Authors: | Wang, A.Y, Schulze, J.M, Skordalakes, E, Berger, J.M, Rine, J, Kobor, M.S. | | Deposit date: | 2008-12-15 | | Release date: | 2009-10-27 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
9BOL
 
 | | Crystal structure of the complex between VHL, ElonginB, ElonginC, and compound 5 | | Descriptor: | (4R)-1-[(2S)-2-(4-cyclopropyl-1H-1,2,3-triazol-1-yl)-3,3-dimethylbutanoyl]-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Murray, J.M, Wu, H, Fuhrmann, J, Fairbrother, W.J, DiPasquale, A. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potency-enhanced peptidomimetic VHL ligands with improved oral bioavailability
To Be Published
|
|
1T27
 
 | | THE STRUCTURE OF PITP COMPLEXED TO PHOSPHATIDYLCHOLINE | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylinositol transfer protein alpha isoform | | Authors: | Yoder, M.D, Thomas, L.M, Tremblay, J.M, Oliver, R.L, Yarbrough, L.R, Helmkamp Jr, G.M. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STRUCTURE OF A MULTIFUNCTIONAL PROTEIN. MAMMALIAN PHOSPHATIDYLINOSITOL TRANSFER PROTEIN COMPLEXED WITH PHOSPHATIDYLCHOLINE
J.Biol.Chem., 276, 2001
|
|
1TEY
 
 | | NMR structure of human histone chaperone, ASF1A | | Descriptor: | ASF1 anti-silencing function 1 homolog A | | Authors: | Mousson, F, Lautrette, A, Thuret, J.Y, Agez, M, Amigues, B, Courbeyrette, R, Neumann, J.M, Guerois, R, Mann, C, Ochsenbein, F. | | Deposit date: | 2004-05-26 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of Asf1 with histone H3 and its functional implications.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TF3
 
 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
1TME
 
 | | THREE-DIMENSIONAL STRUCTURE OF THEILER VIRUS | | Descriptor: | THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP1), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP2), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP3), ... | | Authors: | Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1992-01-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of Theiler virus.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
5U03
 
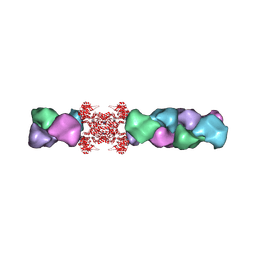 | | Cryo-EM structure of the human CTP synthase filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase 1, URIDINE 5'-TRIPHOSPHATE | | Authors: | Lynch, E.M, Kollman, J.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-04-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5U3C
 
 | |
5U6R
 
 | |
7TQV
 
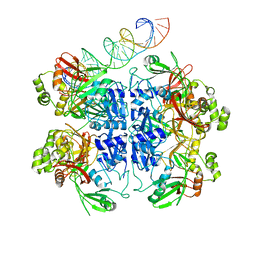 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (33-MER), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
7TJ2
 
 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (31-MER), Uridylate-specific endoribonuclease nsp15 | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
8AF2
 
 | | Human Sterol Carrier Protein with unnatural amino acid 2,2'-bipyridine alanine incorporated at position 111 | | Descriptor: | COPPER (II) ION, Enoyl-CoA hydratase 2, FRAGMENT OF TRITON X-100, ... | | Authors: | Richardson, J.M, Klemencic, E, Jarvis, A.G. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Using BpyAla to generate copper artificial metalloenzymes: a catalytic and structural study.
Catalysis Science And Technology, 14, 2024
|
|
1K5K
 
 | | Homonuclear 1H Nuclear Magnetic Resonance Assignment and Structural Characterization of HIV-1 Tat Mal Protein | | Descriptor: | TAT protein | | Authors: | Gregoire, C, Peloponese, J.M, Esquieu, D, Opi, S, Campbell, G, Solomiac, M, Lebrun, E, Lebreton, J, Loret, E.P. | | Deposit date: | 2001-10-11 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Homonuclear (1)H-NMR assignment and structural characterization of human immunodeficiency virus type 1 Tat Mal protein.
Biopolymers, 62, 2001
|
|
1D6H
 
 | | CHALONE SYNTHASE (N336A MUTANT COMPLEXED WITH COA) | | Descriptor: | CHALCONE SYNTHASE, COENZYME A, SULFATE ION | | Authors: | Jez, J.M, Ferrer, J.L, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1999-10-13 | | Release date: | 2000-02-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
Biochemistry, 39, 2000
|
|
4M1W
 
 | | Crystal Structure of small molecule vinylsulfonamide covalently bound to K-Ras G12C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, MAGNESIUM ION, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4MX5
 
 | | Structure of ricin A chain bound with benzyl-(2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)ethyl)carbamate | | Descriptor: | Ricin A chain, benzyl (2-{[(2-amino-4-oxo-3,4-dihydropteridin-7-yl)carbonyl]amino}ethyl)carbamate | | Authors: | Jasheway, K.R, Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MN4
 
 | | Structural Basis for the MukB-topoisomerase IV Interaction | | Descriptor: | Chromosome partition protein MukB, DNA topoisomerase 4 subunit A | | Authors: | Vos, S.M, Stewart, N.K, Oakley, M.G, Berger, J.M. | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo.
Embo J., 32, 2013
|
|
4N48
 
 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with capped RNA fragment | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
