5I76
 
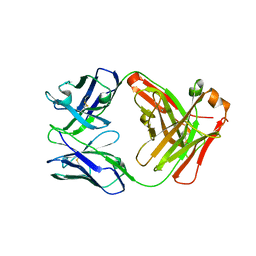 | | Crystal structure of FM318, a recombinant Fab adopted from cetuximab | | Descriptor: | FM318_heavy_cahin, FM318_light_chain | | Authors: | Sim, D.W, Kim, J.H, Seok, S.H, Seo, M.D, Kim, Y.P, Won, H.S. | | Deposit date: | 2016-02-16 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Bacterial production and structure-functional validation of a recombinant antigen-binding fragment (Fab) of an anti-cancer therapeutic antibody targeting epidermal growth factor receptor.
Appl.Microbiol.Biotechnol., 100, 2016
|
|
5I83
 
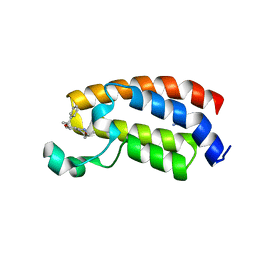 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02773986 | | Descriptor: | (4R)-4-methyl-7-[(1R)-1-phenylethoxy]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, CREB-binding protein, THIOCYANATE ION | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I8E
 
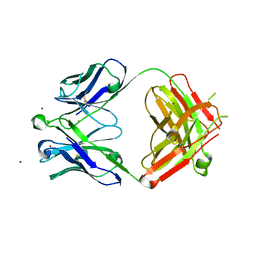 | | Crystal Structure of Broadly Neutralizing HIV-1 Fusion Peptide-Targeting Antibody VRC34.01 Fab | | Descriptor: | VRC34.01 Fab heavy chain, VRC34.01 Fab light chain, ZINC ION | | Authors: | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
2W08
 
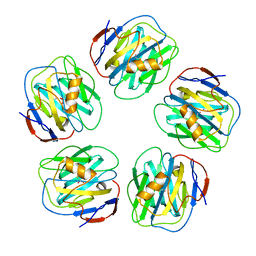 | | The structure of serum amyloid P component bound to 0-phospho- threonine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHOTHREONINE, ... | | Authors: | Kolstoe, S.E, Pepys, M.B, Wood, S.P. | | Deposit date: | 2008-08-12 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Dissection of Alzheimer'S Disease Neuropathology by Depletion of Serum Amyloid P Component.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1JCU
 
 | |
5HL3
 
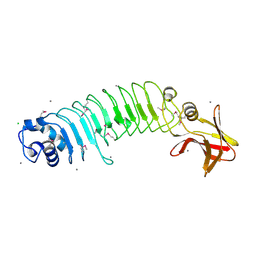 | | Crystal structure of Listeria monocytogenes InlP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lmo2470 protein | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Kiryukhina, O, Kwon, K, Faralla, C, Bakardjiev, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing.
Plos Pathog., 14, 2018
|
|
5HVL
 
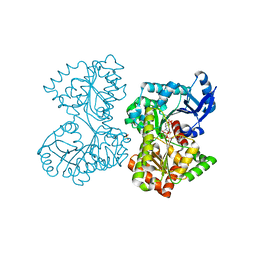 | | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase [UDP-forming], SULFATE ION, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
2W3B
 
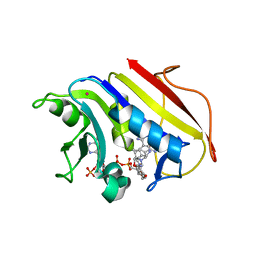 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND A LIPOPHILIC ANTIFOLATE SELECTIVE FOR MYCOBACTERIUM AVIUM DHFR, 6-((2,5- DIETHOXYPHENYL)AMINOMETHYL)-2,4-DIAMINO-5-METHYLPYRIDO(2,3-D) PYRIMIDINE (SRI-8686) | | Descriptor: | 6-{[(2,5-DIETHOXYPHENYL)AMINO]METHYL}-5-METHYLPYRIDO[2,3-D]PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE, FOLIC ACID, ... | | Authors: | Leung, A.K.W, Reynolds, R.C, Borhani, D.W. | | Deposit date: | 2008-11-11 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium Avium Dihydrofolate Reductase by a Lipophilic Antifolate
To be Published
|
|
1JI6
 
 | |
5JDD
 
 | |
1JRM
 
 | |
5JDJ
 
 | | Crystal structure of domain I10 from titin in space group P212121 | | Descriptor: | CALCIUM ION, Titin | | Authors: | Williams, R, Bogomolovas, J, Labiet, S, Mayans, O. | | Deposit date: | 2016-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Exploration of pathomechanisms triggered by a single-nucleotide polymorphism in titin's I-band: the cardiomyopathy-linked mutation T2580I.
Open Biology, 6, 2016
|
|
6UIB
 
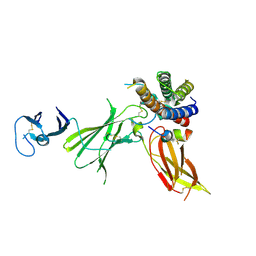 | | Crystal structure of IL23 bound to peptide 23-652 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Durbin, J.D, Wang, J, Afshar, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Integration of phage and yeast display platforms: A reliable and cost effective approach for binning of peptides as displayed on-phage.
Plos One, 15, 2020
|
|
5JNY
 
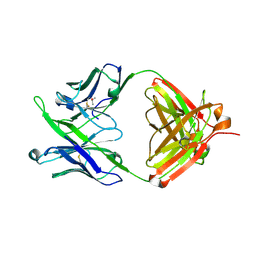 | | Crystal Structure of 10E8 Fab | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, CHLORIDE ION, ... | | Authors: | Ofek, G, Kwong, P. | | Deposit date: | 2016-05-01 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8.
Plos One, 11, 2016
|
|
6WAS
 
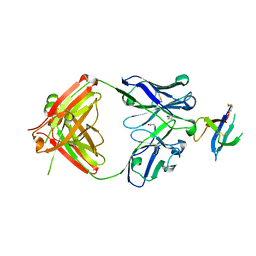 | | Structure of D19.PA8 Fab in complex with 1FD6 16055 V1V2 scaffold | | Descriptor: | 1FD6 16055 V1V2 scaffold, 2-acetamido-2-deoxy-beta-D-glucopyranose, GN1_PA8 Fab Heavy chain, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
2V5U
 
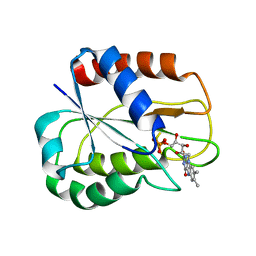 | | I92A FLAVODOXIN FROM ANABAENA | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Martinez-Julvez, M, Herguedas, B, Frago, S, Serrano, A, Molina, R, Hamiaux, C, Schierbeek, B, Medina, M, Hermoso, J.A. | | Deposit date: | 2007-07-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Tuning of the Fmn Binding and Oxido-Reduction Properties by Neighboring Side Chains in Anabaena Flavodoxin.
Arch.Biochem.Biophys., 467, 2007
|
|
5J17
 
 | |
5JRS
 
 | |
5JOE
 
 | | Crystal structure of I81 from titin | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Titin | | Authors: | Fleming, J, Zhou, T, Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Febs Lett., 590, 2016
|
|
5K6V
 
 | | Sidekick-1 immunoglobulin domains 1-4, crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein sidekick-1, ... | | Authors: | Jin, X, Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.208 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
2UZC
 
 | | Structure of human PDLIM5 in complex with the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ AND LIM DOMAIN 5 | | Authors: | Bunkoczi, G, Elkins, J, Salah, E, Burgess-Brown, N, Papagrigoriou, E, Pike, A.C.W, Turnbull, A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D. | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms.
Protein Sci., 19, 2010
|
|
6VTT
 
 | |
6VWS
 
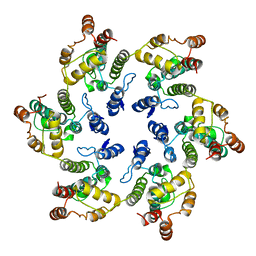 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
5IPJ
 
 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolopyrrolone inhibitor. | | Descriptor: | 2-(tert-butylamino)-3-methyl-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of Quinazolinone-pyrrolopyrrolones as Potent and Orally Bioavailable Pan-Pim Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5JL4
 
 | |
