1MT6
 
 | | Structure of histone H3 K4-specific methyltransferase SET7/9 with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET9 | | Authors: | Jacobs, S.A, Harp, J.M, Devarakonda, S, Kim, Y, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active site of the SET domain is constructed on a knot
Nat.Struct.Biol., 9, 2002
|
|
2HBR
 
 | |
2HBZ
 
 | |
2HBY
 
 | |
2HCB
 
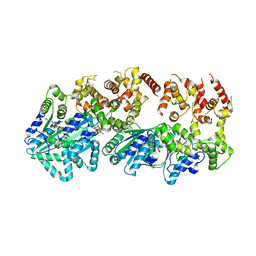 | | Structure of AMPPCP-bound DnaA from Aquifex aeolicus | | Descriptor: | Chromosomal replication initiator protein dnaA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Erzberger, J.P, Mott, M.L, Berger, J.M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2HCO
 
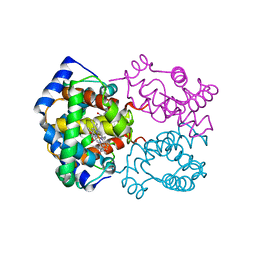 | |
1N3K
 
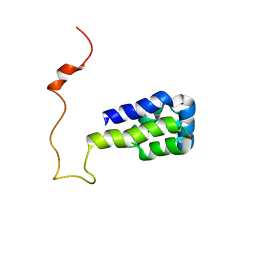 | | Solution structure of phosphoprotein enriched in astrocytes 15 kDa (PEA-15) | | Descriptor: | Astrocytic phosphoprotein PEA-15 | | Authors: | Hill, J.M, Vaidyanathan, H, Ramos, J.W, Ginsberg, M.H, Werner, M.H. | | Deposit date: | 2002-10-28 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of ERK MAP Kinase by PEA-15 Reveals a Common Docking Site Within the Death Domain and Death Effector Domain
Embo J., 21, 2002
|
|
1NE4
 
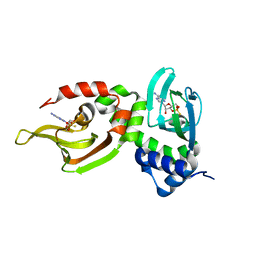 | | Crystal Structure of Rp-cAMP Binding R1a Subunit of cAMP-dependent Protein Kinase | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Jones, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2002-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of RIalpha Subunit of Cyclic Adenosine 5'-Monophosphate (cAMP)-Dependent Protein Kinase Complexed with (R(p))-Adenosine 3',5'-Cyclic Monophosphothioate and (S(p))-Adenosine 3',5'-Cyclic Monophosphothioate, the Phosphothioate Analogues of cAMP.
Biochemistry, 43, 2004
|
|
1MTX
 
 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF MARGATOXIN BY 1H, 13C, 15N TRIPLE-RESONANCE NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MARGATOXIN | | Authors: | Johnson, B.A, Stevens, S.P, Williamson, J.M. | | Deposit date: | 1994-12-27 | | Release date: | 1995-11-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of margatoxin by 1H, 13C, 15N triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1NCV
 
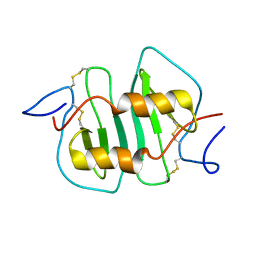 | | DETERMINATION CC-CHEMOKINE MCP-3, NMR, 7 STRUCTURES | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 3 | | Authors: | Meunier, S, Bernassau, J.M, Guillemot, J.C, Ferrara, P, Darbon, H. | | Deposit date: | 1997-02-05 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of CC chemokine monocyte chemoattractant protein 3 by 1H two-dimensional NMR spectroscopy.
Biochemistry, 36, 1997
|
|
2FNE
 
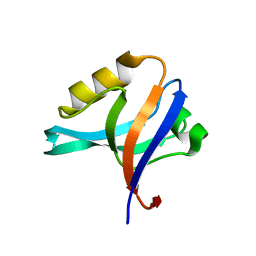 | | The crystal structure of the 13th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Berridge, G, Johansson, C, Colebrook, S, Salah, E, Burgess, N, Smee, C, Savitsky, P, Bray, J, Schoch, G, Phillips, C, Gileadi, C, Soundarajan, M, Yang, X, Elkins, J.M, Gorrec, F, Turnbull, A, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
1N91
 
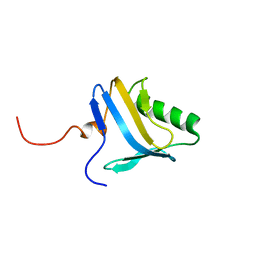 | | Solution NMR Structure of Protein yggU from Escherichia coli. Northeast Structural Genomics Consortium Target ER14. | | Descriptor: | orf, hypothetical protein | | Authors: | Aramini, J.M, Xiao, R, Huang, Y.J, Acton, T.B, Wu, M.J, Mills, J.L, Tejero, R.T, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-11-21 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Resonance assignments for the hypothetical protein yggU from Escherichia coli.
J.Biomol.Nmr, 27, 2003
|
|
1NAT
 
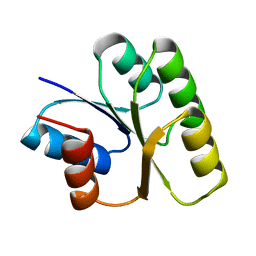 | | CRYSTAL STRUCTURE OF SPOOF FROM BACILLUS SUBTILIS | | Descriptor: | SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Zapf, J, Hoch, J.A, Whiteley, J.M, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: crystal structure of Spo0F from Bacillus subtilis.
Biochemistry, 36, 1997
|
|
2FQQ
 
 | | Crystal structure of human caspase-1 (Cys285->Ala, Cys362->Ala, Cys364->Ala, Cys397->Ala) in complex with 1-methyl-3-trifluoromethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid (2-mercapto-ethyl)-amide | | Descriptor: | 1-METHYL-3-TRIFLUOROMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID (2-MERCAPTO-ETHYL)-AMIDE, Caspase-1 | | Authors: | Scheer, J.M, Wells, J.A, Romanowski, M.J. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A common allosteric site and mechanism in caspases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FMQ
 
 | | Sodium in active site of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FO0
 
 | | Organization of the SH3-SH2 Unit in Active and Inactive Forms of the c-Abl Tyrosine Kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Seeliger, M, Davies, J.M, Weis, W.I, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase.
Mol.Cell, 21, 2006
|
|
1O26
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and dUMP at 1.6 A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, TRIETHYLENE GLYCOL, ... | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1NXJ
 
 | | Structure of Rv3853 from Mycobacterium tuberculosis | | Descriptor: | GLYOXYLIC ACID, L(+)-TARTARIC ACID, Probable S-adenosylmethionine:2-demethylmenaquinone methyltransferase | | Authors: | Johnston, J.M, Arcus, V.L, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-10 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Putative Methyltransferase from Mycobacterium tuberculosis: Misannotation of a Genome Clarified by Protein Structural Analysis
J.Bacteriol., 185, 2003
|
|
1O2B
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and PO4 at 2.45 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
2IHN
 
 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
2IEN
 
 | | Crystal structure analysis of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
1O25
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with dUMP at 2.4 A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
2IF6
 
 | | Crystal structure of metalloprotein yiiX from Escherichia coli O157:H7, DUF1105 | | Descriptor: | Hypothetical protein yiiX, UNKNOWN ATOM OR ION | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical metalloprotein yiiX from Escherichia coli O157:H7
To be Published
|
|
2ISQ
 
 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Arabidopsis Thaliana in Complex with C-Terminal Peptide from Arabidopsis Serine Acetyltransferase | | Descriptor: | Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Francois, J.A, Kumaran, S, Jez, J.M. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for interaction of o-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex.
Plant Cell, 18, 2006
|
|
2IEO
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Protease, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
