6JV2
 
 | | Structure of RyR2 (P/L-Ca2+/Ca2+-CaM dataset) | | Descriptor: | CALCIUM ION, Calmodulin-1, RyR2, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
3OCI
 
 | | Crystal structure of TBP (TATA box binding protein) | | Descriptor: | 1,2-ETHANEDIOL, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Cui, S, Wollmann, P, Moldt, M, Hopfner, K.-P. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
6KNW
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
2O2U
 
 | | Crystal structure of human JNK3 complexed with N-(3-cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)-2-fluorobenzamide | | Descriptor: | Mitogen-activated protein kinase 10, N-(3-cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)-2-fluorobenzamide | | Authors: | Somers, D, Rowland, P. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6KNV
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
7CDA
 
 | | Crystal structure of T2R-TTL-PAC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.659 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
7CE8
 
 | | Crystal structure of T2R-TTL-Compound11 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
7CEK
 
 | | Crystal structure of T2R-TTL-BML-284 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
7CE6
 
 | | Crystal structure of T2R-TTL-Compound9 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
1GSG
 
 | | Structure of E.coli glutaminyl-tRNA synthetase complexed with trnagln and ATP at 2.8 Angstroms resolution | | Descriptor: | GLUTAMINYL-TRNA SYNTHETASE, TRNAGLN | | Authors: | Rould, M.A, Perona, J.J, Soell, D, Steitz, T.A. | | Deposit date: | 1990-04-03 | | Release date: | 1992-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution.
Science, 246, 1989
|
|
5X74
 
 | | The crystal Structure PDE delta in complex with compound (R, R)-1g | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-[2-[4-[(2R)-2-(2-fluorophenyl)-4-oxidanylidene-1,2-dihydroquinazolin-3-yl]piperidin-1-yl]ethyl]-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5X73
 
 | | The crystal Structure PDE delta in complex with R-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5TDK
 
 | | RNA decamer duplex with eight 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
5X72
 
 | | The crystal Structure PDE delta in complex with (rac)-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, (2S)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5TKO
 
 | | RNA heptamer duplex with one 2'-5'-linkage | | Descriptor: | COBALT (II) ION, RNA (5'-R(*GP*GP*AP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-10-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | RNA heptamer duplex with one 2'-5'-linkage
To Be Published
|
|
5TDJ
 
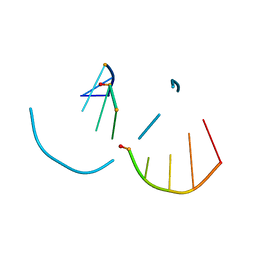 | | RNA decamer duplex with four 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
6S41
 
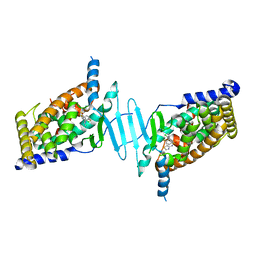 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH XPC-7455 | | Descriptor: | 4-[[(1~{S})-1-[2,5-bis(fluoranyl)phenyl]ethyl]amino]-5-chloranyl-2-fluoranyl-~{N}-(1,3-thiazol-4-yl)benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Focken, T, Maskos, K, Griessner, A, Krapp, S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of CNS-Penetrant Aryl Sulfonamides as Isoform-Selective NaV1.6 Inhibitors with Efficacy in Mouse Models of Epilepsy.
J.Med.Chem., 62, 2019
|
|
5THO
 
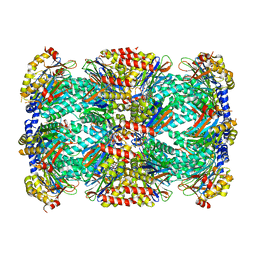 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome in complex with N,C-capped Dipeptide Inhibitor PKS2205 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2016-09-30 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
7BPB
 
 | | Human AAA+ ATPase VCP mutant - T76E, AMP-PNP bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-22 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BP9
 
 | | Human AAA+ ATPase VCP mutant - T76E, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BP8
 
 | | Human AAA+ ATPase VCP mutant - T76A, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BPA
 
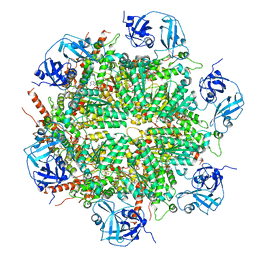 | | Human AAA+ ATPase VCP mutant - T76A, AMP-PNP-bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
5U0Q
 
 | | RNA-DNA heptamer duplex with one 2'-5'-linkage | | Descriptor: | COBALT (II) ION, DNA/RNA (5'-R(*GP*GP*AP*GP*C)-D(P*T)-R(P*A)-3'), MAGNESIUM ION, ... | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-11-26 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | RNA-DNA heptamer duplex with one 2'-5'-linkage
To Be Published
|
|
4HK5
 
 | | Crystal structure of Cordyceps militaris IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4HK7
 
 | | Crystal structure of Cordyceps militaris IDCase in complex with uracil | | Descriptor: | URACIL, Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
