7Q2J
 
 | | Quaternary Complex of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC Homer | | Descriptor: | Elongin-B, Elongin-C, N-[5-[4-[[5-[[(2S)-3,3-dimethyl-1-[(2S,4R)-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-5-oxidanylidene-pentyl]carbamoyl]phenyl]-2-(4-methylpiperazin-1-yl)phenyl]-6-oxidanylidene-4-(trifluoromethyl)-1H-pyridine-3-carboxamide, ... | | Authors: | Kraemer, A, Doelle, A, Schwalm, M.P, Adhikari, B, Wolf, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
3NIZ
 
 | | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with ADP bound. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rhodanese family protein | | Authors: | Wernimont, A.K, Dong, A, Lew, J, Lin, Y.H, Hassanali, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with ADP bound.
To be Published
|
|
8QWK
 
 | | Structure of p53 cancer mutant Y126C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Markl, A.M, Balourdas, D.I, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWP
 
 | | Structure of p53 cancer mutant Y236C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWO
 
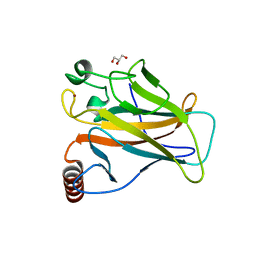 | | Structure of p53 cancer mutant Y234C | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWM
 
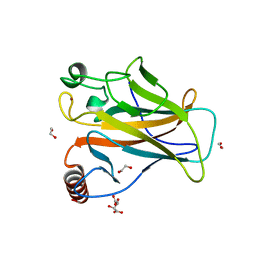 | | Structure of p53 cancer mutant Y205C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWN
 
 | | Structure of p53 cancer mutant Y220C (hexagonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWL
 
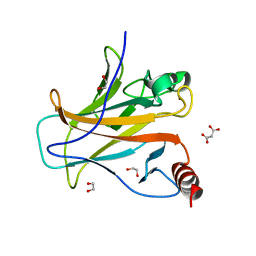 | | Structure of p53 cancer mutant Y163C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, MALONATE ION, ... | | Authors: | Balourdas, D.I, Markl, A.M, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
7B6M
 
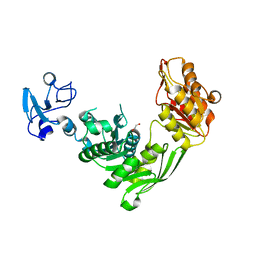 | | Crystal structure of MurE from E.coli in complex with Z1198948504 | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(2,5-Dioxopyrrolidin-1-yl)ethyl]-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC), Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7P7F
 
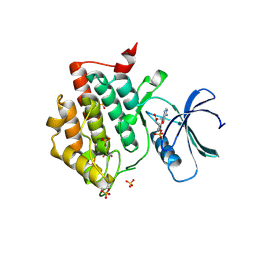 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
7P7G
 
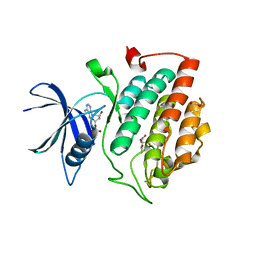 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
7P7H
 
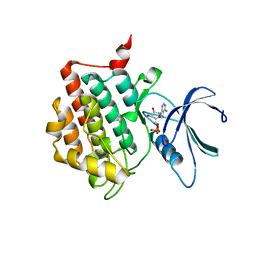 | |
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
7ORF
 
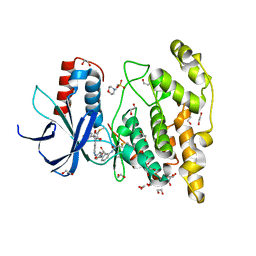 | | Crystal structure of JNK3 in complex with FMU-001-367 (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 10, ... | | Authors: | Chaikuad, A, Koch, P, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8BZI
 
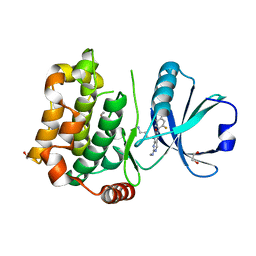 | | Human MST3 (STK24) kinase in complex with inhibitor MR39 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2,5-bis(fluoranyl)-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
7ORE
 
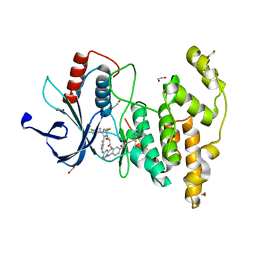 | | Crystal structure of JNK3 in complex with light-activated covalent inhibitor MR-II-249 with both non-covalent and covalent binding modes (compound 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[(5Z)-9-[[4-[5-(4-fluorophenyl)-3-methyl-2-methylsulfanyl-imidazol-4-yl]pyridin-2-yl]amino]-11,12-dihydrobenzo[c][1,2]benzodiazocin-2-yl]butanamide, Mitogen-activated protein kinase 10 | | Authors: | Chaikuad, A, Reynders, M, Trauner, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OPS
 
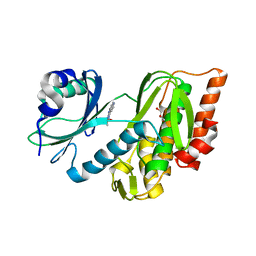 | | Crystal structure of haspin in complex with ZW282 (compound 2a) | | Descriptor: | 2-methylsulfanyl-10-nitro-pyrido[3,4-g]quinazoline, GLYCEROL, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Anizon, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synthesis and biological evaluation of Haspin inhibitors: Kinase inhibitory potency and cellular activity.
Eur.J.Med.Chem., 236, 2022
|
|
5B8D
 
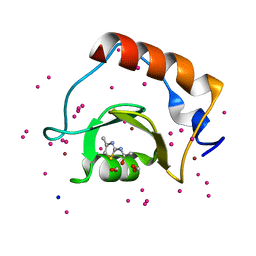 | | Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | FORMIC ACID, Histone deacetylase 6, SODIUM ION, ... | | Authors: | Harding, R.J, Tempel, W, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Ravichandran, M, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
5C8G
 
 | | Crystal Structure Analysis of PP-BRD20 from Tb427tmp complexed with BI-2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, PP-BRD20, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, Hou, C.F.D, El Bakkouri, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Amain, M, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Analysis of PP-BRD20 from Tb427tmp
to be published
|
|
7OR1
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
TO BE PUBLISHED
|
|
7P4E
 
 | | Crystal structure of PPARgamma in complex with compound FL217 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma, SULFATE ION, ... | | Authors: | Ni, X, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7P4K
 
 | | Soluble epoxide hydrolase in complex with FL217 | | Descriptor: | Bifunctional epoxide hydrolase 2, ~{N}-[[4-(cyclopropylsulfonylamino)-2-(trifluoromethyl)phenyl]methyl]-1-[(3-fluorophenyl)methyl]indole-5-carboxamide | | Authors: | Ni, X, Kramer, J.S, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
4LZ2
 
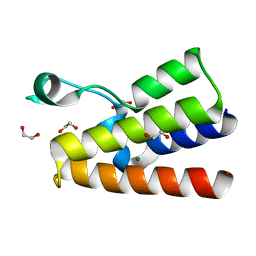 | | Crystal structure of the bromodomain of human BAZ2A | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2A, MAGNESIUM ION | | Authors: | Tallant, C, Nunez-Alonso, G, Picaud, S, Filippakopoulos, P, Krojer, T, Bradley, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
8BZJ
 
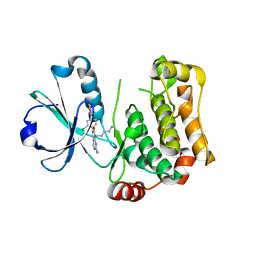 | | Human MST3 (STK24) kinase in complex with inhibitor MRLW5 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8C7Y
 
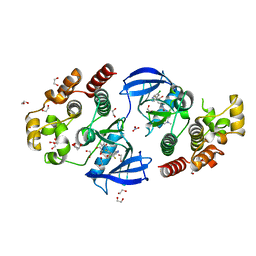 | | Crystal structure of BRAF V600E in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
