4O8I
 
 | | 1.45A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
1SMP
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN SERRATIA MARCESCENS METALLO-PROTEASE AND AN INHIBITOR FROM ERWINIA CHRYSANTHEMI | | Descriptor: | CALCIUM ION, ERWINIA CHRYSANTHEMI INHIBITOR, SERRATIA METALLO PROTEINASE, ... | | Authors: | Baumann, U, Bauer, M, Letoffe, S, Delepelaire, P, Wandersman, C. | | Deposit date: | 1995-01-13 | | Release date: | 1996-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a complex between Serratia marcescens metallo-protease and an inhibitor from Erwinia chrysanthemi.
J.Mol.Biol., 248, 1995
|
|
4OR2
 
 | | Human class C G protein-coupled metabotropic glutamate receptor 1 in complex with a negative allosteric modulator | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{4-[6-(propan-2-ylamino)pyrimidin-4-yl]-1,3-thiazol-2-yl}benzamide, CHOLESTEROL, ... | | Authors: | Wu, H, Wang, C, Gregory, K.J, Han, G.W, Cho, H.P, Xia, Y, Niswender, C.M, Katritch, V, Cherezov, V, Conn, P.J, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator
Science, 344, 2014
|
|
4N40
 
 | | Crystal structure of human Epithelial cell-transforming sequence 2 protein | | Descriptor: | Protein ECT2 | | Authors: | Zou, Y, Shao, Z.H, Li, F.D, Gong, D, Wang, C, Gong, Q, Shi, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Crystal structure of triple-BRCT-domain of ECT2 and insights into the binding characteristics to CYK-4
Febs Lett., 588, 2014
|
|
7BYJ
 
 | | Crystal structure of the FERM domain of FRMPD4 | | Descriptor: | FERM and PDZ domain-containing protein 4 | | Authors: | Lin, L, Wang, M, Wang, C, Zhu, J. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the FERM domain of a neural scaffold protein FRMPD4 implicated in X-linked intellectual disability.
Biochem.J., 477, 2020
|
|
8W8D
 
 | |
8IN0
 
 | |
2MY5
 
 | | Solution Structure of KstB-PCP in kosinostatin biosynthesis | | Descriptor: | N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(2-sulfanylethyl)-beta-alaninamide, Peptidyl carrier protein | | Authors: | Zhao, B, Lan, W, Wang, C, Tang, G, Cao, C. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N Assigned Chemical Shifts for KstB-PCP
To be Published
|
|
2MY6
 
 | |
2MVT
 
 | | Solution structure of scoloptoxin SSD609 from Scolopendra mutilans | | Descriptor: | Scoloptoxin SSD609 | | Authors: | Wu, F, Sun, P, Wang, C, He, Y, Zhang, L, Tian, C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit.
Sci Rep, 5, 2015
|
|
6K2H
 
 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
7CE1
 
 | | Complex STRUCTURE OF TRANSCRIPTION FACTOR SghR with its COGNATE DNA | | Descriptor: | LacI-type transcription factor, promoter DNA | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7CDV
 
 | | STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR | | Descriptor: | LacI-type transcription factor | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7CDX
 
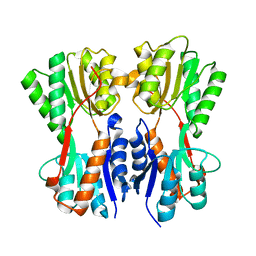 | | Complex STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR with its effector sucrose | | Descriptor: | LacI-type transcription factor, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
6L94
 
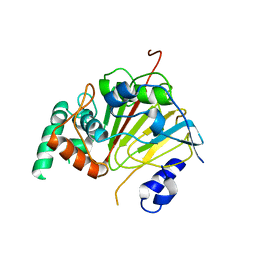 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
6KZJ
 
 | | Crystal structure of Ankyrin B/NdeL1 complex | | Descriptor: | Ankyrin-2, Nuclear distribution protein nudE-like 1 | | Authors: | Ye, J, Li, J, Ye, F, Zhang, M, Zhang, Y, Wang, C. | | Deposit date: | 2019-09-24 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into the interactions of dynein regulator Ndel1 with neuronal ankyrins and implications in polarity maintenance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LOD
 
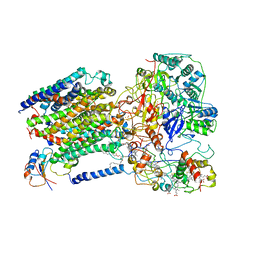 | | Cryo-EM structure of the air-oxidized photosynthetic alternative complex III from Roseiflexus castenholzii | | Descriptor: | Cytochrome c domain-containing protein, FE3-S4 CLUSTER, Fe-S-cluster-containing hydrogenase components 1-like protein, ... | | Authors: | Shi, Y, Xin, Y.Y, Wang, C, Blankenship, R.E, Sun, F, Xu, X.L. | | Deposit date: | 2020-01-05 | | Release date: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the air-oxidized and dithionite-reduced photosynthetic alternative complex III from Roseiflexus castenholzii .
Sci Adv, 6, 2020
|
|
6MRO
 
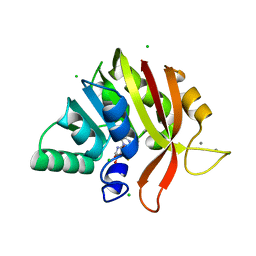 | | Crystal structure of methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution, Northeast Structural Genomics Consortium (NESG) Target MvR53. | | Descriptor: | CALCIUM ION, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Forouhar, F, Wang, C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-10-15 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution.
To Be Published
|
|
6MUU
 
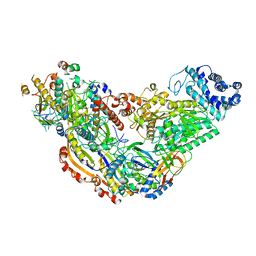 | | Cryo-EM structure of Csm-crRNA binary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (25-MER), Uncharacterized protein Csm1, Uncharacterized protein Csm2, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6N9N
 
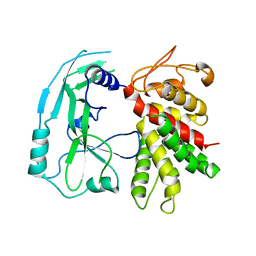 | | Crystal structure of murine GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
6N9O
 
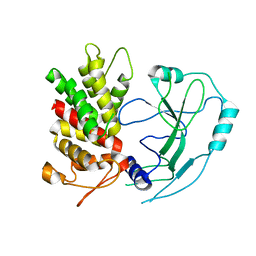 | | Crystal structure of human GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
6MUR
 
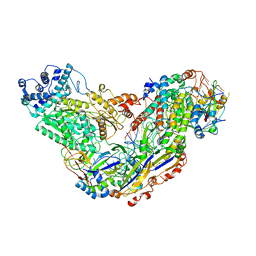 | | Cryo-EM structure of Csm-crRNA-target RNA ternary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (27-MER), RNA (5'-R(P*GP*CP*AP*AP*AP*CP*CP*GP*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*C)-3'), Uncharacterized protein, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
7ECA
 
 | |
6MUT
 
 | | Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*GP*C)-3'), RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP*C)-3'), Uncharacterized protein Csm1, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
