7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | Descriptor: | ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | Descriptor: | bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
6KVF
 
 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | Descriptor: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
8HOG
 
 | | Crystal structure of Bcl-2 in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOH
 
 | | Crystal structure of Bcl-2 G101V in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOI
 
 | | Crystal structure of Bcl-2 D103Y in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, FORMIC ACID, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
6U3U
 
 | | Crystal Structure of Shiga Toxin 2K | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Shiga toxin 2K subunit A, ... | | Authors: | Zhang, Y.Z, He, X.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2.
Microorganisms, 8, 2019
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
6LPC
 
 | | Crystal Structure of rat Munc18-1 with K332E/K333E mutation | | Descriptor: | Syntaxin-binding protein 1 | | Authors: | Wang, X.P, Gong, J.H, Wang, S, Zhu, L, Yang, X.Y, Xu, Y.Y, Yang, X.F, Ma, C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | Munc13 activates the Munc18-1/syntaxin-1 complex and enables Munc18-1 to prime SNARE assembly.
Embo J., 39, 2020
|
|
8HOM
 
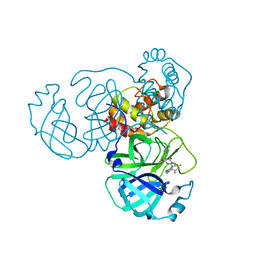 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
8HOL
 
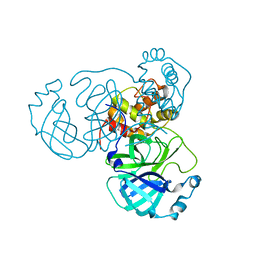 | |
8HOZ
 
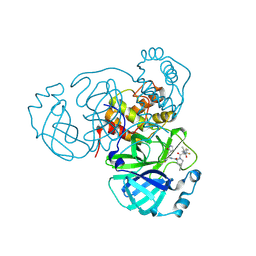 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
6U4Y
 
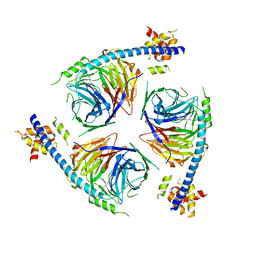 | | Crystal Structure of an EZH2-EED Complex in an Oligomeric State | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A partially disordered region connects gene repression and activation functions of EZH2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
9JAL
 
 | | Cryo-EM structure of MPXV core protease in complex with compound A1 | | Descriptor: | A1ECM-DI8-ALA-ETA, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9JAN
 
 | | Cryo-EM structure of MPXV protease in complex with compound A4 | | Descriptor: | A1ECL-DI8-ALA-AEM, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9JAM
 
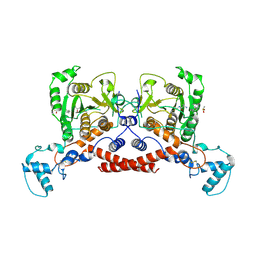 | | Cryo-EM structure of MPXV core protease in complex with compound A3 | | Descriptor: | A1ECK-DI8-ALA-AEM, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9JAQ
 
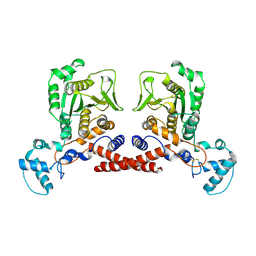 | | Cryo-EM structure of MPXV core protease in the apo-form | | Descriptor: | Core protease I7 | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9KQV
 
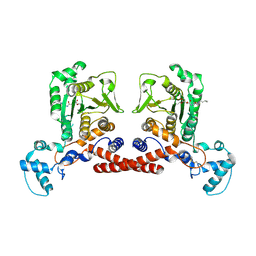 | | Cryo-EM structure of MPXV core protease in complex with aloxistatin(E64d) | | Descriptor: | Core protease I7, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9KR6
 
 | | Cryo-EM structure of MPXV core protease in complex with the substrate derivative I-G18 | | Descriptor: | Core protease I7, Core protein VP8 | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3DIW
 
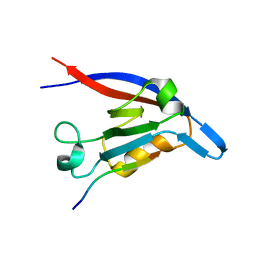 | | c-terminal beta-catenin bound TIP-1 structure | | Descriptor: | Tax1-binding protein 3, decameric peptide form Catenin beta-1 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
3DJ1
 
 | | crystal structure of TIP-1 wild type | | Descriptor: | SULFATE ION, Tax1-binding protein 3 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1.
J.Mol.Biol., 384, 2008
|
|
8HTS
 
 | | Crystal structure of Bcl2 in complex with S-10r | | Descriptor: | 4-[2-[(2~{S})-2-(2-cyclopropylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
