1DXW
 
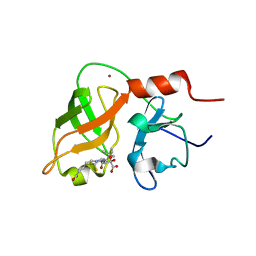 | | structure of hetero complex of non structural protein (NS) of hepatitis C virus (HCV) and synthetic peptidic compound | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Cordier, F, Narjes, F, Gerlach, B, Sambucini, S, Grzesiek, S, Matassa, V.G, Defrancesco, R, Bazzo, R. | | Deposit date: | 2000-01-17 | | Release date: | 2001-01-12 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Inhibitor Binding Induces Active Site Stabilisation of the Hcv Ns3 Protein Serine Protease Domain
Embo J., 19, 2000
|
|
3ADK
 
 | |
2PNL
 
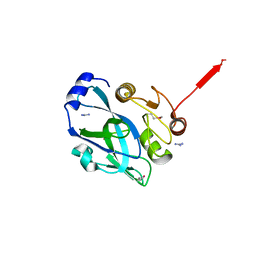 | | Crystal structure of VP4 protease from infectious pancreatic necrosis virus (IPNV) in space group P1 | | Descriptor: | GUANIDINE, Protease VP4 | | Authors: | Paetzel, M, Lee, J, Feldman, A.R, Delmas, B. | | Deposit date: | 2007-04-24 | | Release date: | 2007-06-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the VP4 protease from infectious pancreatic necrosis virus reveals the acyl-enzyme complex for an intermolecular self-cleavage reaction.
J.Biol.Chem., 282, 2007
|
|
2PNM
 
 | |
7T5S
 
 | | P. aeruginosa LpxA in complex with ligand H16 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, N~2~-(cyclohexylacetyl)-N-1H-tetrazol-5-yl-L-alaninamide | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5R
 
 | | P. aeruginosa LpxA in complex with ligand H7 | | Descriptor: | 3-bromo-N-[3-(1H-tetrazol-5-yl)phenyl]-1H-indole-5-carboxamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5X
 
 | | P. aeruginosa LpxA in complex with ligand L6 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, Nalpha-(tert-butoxycarbonyl)-N-1H-tetrazol-5-yl-D-tryptophanamide | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5Z
 
 | | P. aeruginosa LpxA in complex with ligand L8 | | Descriptor: | (4S)-N-(1H-tetrazol-5-yl)-2-[3-(trifluoromethyl)benzene-1-sulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T60
 
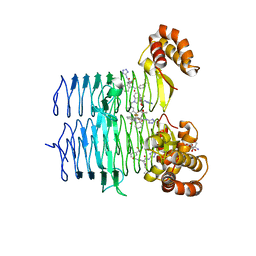 | | P. aeruginosa LpxA in complex with ligand L13 | | Descriptor: | (3S)-3-(5,5-dimethyl-2-oxo-1,3-oxazolidin-3-yl)-N-(1H-tetrazol-5-yl)-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T61
 
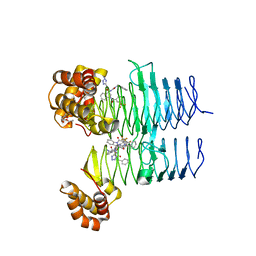 | | P. aeruginosa LpxA in complex with ligand L15 | | Descriptor: | (3S)-3-({[(Z)-phenylmethylidene]carbamoyl}amino)-N-(1H-tetrazol-5-yl)-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indole-3-carboxamide, ACETATE ION, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
5FPO
 
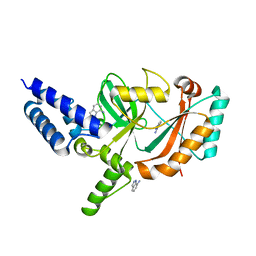 | | Structure of Bacterial DNA Ligase with small-molecule ligand 1H- indazol-7-amine (AT4213) in an alternate binding site. | | Descriptor: | 1H-indazol-7-amine, DNA LIGASE | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPR
 
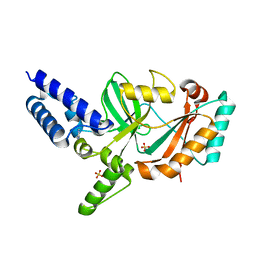 | | Structure of Bacterial DNA Ligase with small-molecule ligand pyrimidin-2-amine (AT371) in an alternate binding site. | | Descriptor: | DNA LIGASE, PYRIMIDIN-2-AMINE, SULFATE ION | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7UVF
 
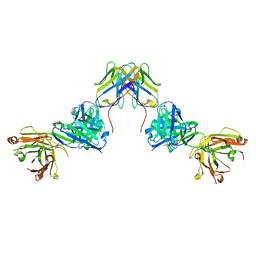 | | Crystal structure of ZED8 Fab complex with CD8 alpha | | Descriptor: | CHLORIDE ION, GLYCEROL, Immunoglobulin heavy chain, ... | | Authors: | Yu, C, Davies, C, Koerber, J.T, Williams, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Preclinical development of ZED8, an 89 Zr immuno-PET reagent for monitoring tumor CD8 status in patients undergoing cancer immunotherapy.
Eur J Nucl Med Mol Imaging, 50, 2023
|
|
2E9T
 
 | | Foot-and-mouth disease virus RNA-polymerase RNA dependent in complex with a template-primer RNA and 5F-UTP | | Descriptor: | 5'-R(*GP*GP*GP*CP*CP*CP*(5FU))-3', 5'-R(P*UP*AP*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3AUQ
 
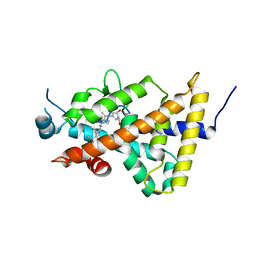 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with Yne-diene type analog of active 14-epi-2alpha-methyl-19-norvitamin D3 | | Descriptor: | (1R,2S,3R)-5-[2-[(1R,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-7a-methyl-1,2,3,3a,6,7-hexahydroinden-4-yl]ethynyl]-2-methyl-cyclohex-4-ene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Development of 14-epi-19-nortachysterol and its unprecedented binding configuration for the human vitamin D receptor
J.Am.Chem.Soc., 133, 2011
|
|
3AUR
 
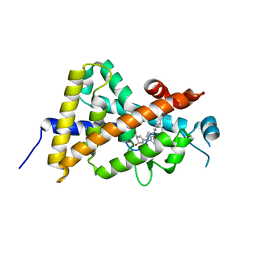 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with Yne-diene type analog of active 14-epi-2beta-methyl-19-norvitamin D3 | | Descriptor: | (1R,2S,3R)-5-[2-[(1R,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-7a-methyl-1,2,3,3a,6,7-hexahydroinden-4-yl]ethynyl]-2-methyl-cyclohex-4-ene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Development of 14-epi-19-nortachysterol and its unprecedented binding configuration for the human vitamin D receptor
J.Am.Chem.Soc., 133, 2011
|
|
7UMM
 
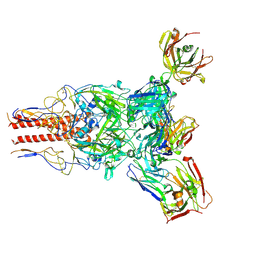 | | H1 Solomon Islands 2006 hemagglutinin in complex with Ab109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Windsor, I.W, Caradonna, T.M, Schmidt, A.G. | | Deposit date: | 2022-04-07 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | An epitope-enriched immunogen expands responses to a conserved viral site.
Cell Rep, 41, 2022
|
|
2FJC
 
 | | Crystal structure of antigen TpF1 from Treponema pallidum | | Descriptor: | Antigen TpF1, FE (III) ION | | Authors: | Thumiger, A, Polenghi, A, Papinutto, E, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2006-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of antigen TpF1 from Treponema pallidum.
Proteins, 62, 2006
|
|
3AX8
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 15alpha-methoxy-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3S,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-3-methoxy-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-03-30 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New C15-substituted active vitamin D3
Org.Lett., 13, 2011
|
|
2E9R
 
 | | Foot-and-mouth disease virus RNA-dependent RNA polymerase in complex with a template-primer RNA and with ribavirin | | Descriptor: | 5'-R(*CP*AP*UP*GP*GP*GP*CP*CP*C)-3', 5'-R(*CP*CP*C*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EC0
 
 | | RNA-dependent RNA polymerase of foot-and-mouth disease virus in complex with a template-primer RNA and ATP | | Descriptor: | 5'-R(*GP*GP*GP*CP*CP*CP*A)-3', 5'-R(P*AP*UP*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3X31
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 7,8-cis-14-epi-1a,25-Dihydroxy-19-norvitamin D3 | | Descriptor: | (1R,3R,7Z,14beta,17alpha)-17-[(2R)-6-hydroxy-6-methylheptan-2-yl]-9,10-secoestra-5,7-diene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2015-01-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Revisiting the 7,8-cis-vitamin D3 derivatives: synthesis, evaluating the biological activity, and study of the binding configuration
To be Published
|
|
3X36
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 7,8-cis-1a,25-Dihydroxy-19-norvitamin D3 | | Descriptor: | (1R,3R)-5-[(2Z)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Takimoto-Kamimura, M, Kakuda, S. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Revisiting the 7,8-cis-vitamin D3 derivatives: synthesis, evaluating the biological activity, and study of the binding configuration
To be Published
|
|
3AKY
 
 | |
3AUN
 
 | | Crystal structure of the rat vitamin D receptor ligand binding domain complexed with YR335 and a synthetic peptide containing the NR2 box of DRIP 205 | | Descriptor: | (2R)-2-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}butane-1,4-diol, DRIP 205 NR2 box peptide, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Design, synthesis and X-ray crystallographic study of new nonsecosteroidal vitamin D receptor ligands
Bioorg.Med.Chem.Lett., 21, 2011
|
|
