3OW9
 
 | |
3FTH
 
 | |
1LGR
 
 | |
3NVE
 
 | |
5KMZ
 
 | |
3NVF
 
 | |
3NVG
 
 | |
1LOJ
 
 | | Crystal structure of a Methanobacterial Sm-like archaeal protein (SmAP1) bound to uridine-5'-monophosphate (UMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, URIDINE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Mura, C, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-05-06 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs)
Protein Sci., 12, 2003
|
|
1M5Q
 
 | | Crystal structure of a novel Sm-like archaeal protein from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, CADMIUM ION, GLYCEROL, ... | | Authors: | Mura, C, Phillips, M, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-07-09 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and assembly of an augmented Sm-like archaeal protein 14-mer
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5CY5
 
 | | Crystal structure of the T33-51H designed self-assembling protein tetrahedron | | Descriptor: | T33-51H-A, T33-51H-B | | Authors: | Cannon, K.A, Cascio, D, Park, R, Boyken, S, King, N, Yeates, T.O. | | Deposit date: | 2015-07-30 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
4IOD
 
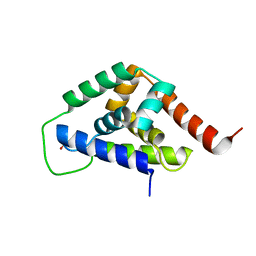 | |
5W50
 
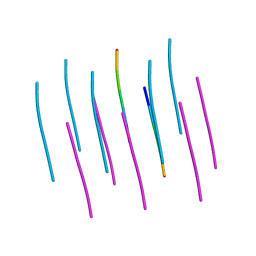 | | Crystal structure of the segment, LIIKGI, from the RRM2 of TDP-43, residues 248-253 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Trinh, H, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-06-13 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic-level evidence for packing and positional amyloid polymorphism by segment from TDP-43 RRM2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WIA
 
 | | Crystal structure of the segment, GNNSYS, from the low complexity domain of TDP-43, residues 370-375 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Trinh, H, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.002 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4IRF
 
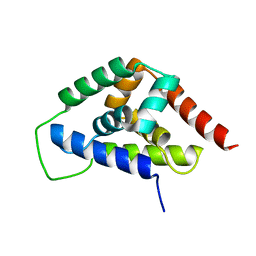 | |
5WHN
 
 | | Crystal structure of the segment, NFGAFS, from the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | Segment of TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-17 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5W7V
 
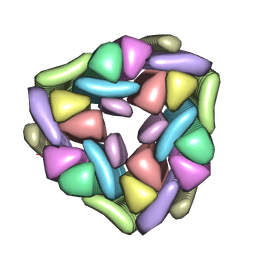 | |
5WHP
 
 | | Crystal structure of the segment, NFGTFS, from the A315T familial variant of the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | Segment of TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WIQ
 
 | | Crystal structure of the segment, GFNGGFG, from the low complexity domain of TDP-43, residues 396-402 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-19 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CF4
 
 | | Segment NFGTFS, with familial mutation A315T and phosphorylated threonine, from the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | NFGTFS | | Authors: | Guenther, E.L, Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.75 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6WLA
 
 | | Antigen binding fragment of ch128.1 | | Descriptor: | Fab ch128.1 heavy chain, Fab ch128.1 light chain, GLYCEROL | | Authors: | Helguera, G, Rodriguez, J.A, Sawaya, M, Cascio, D, Zink, S, Ziegenbein, J, Short, C. | | Deposit date: | 2020-04-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Host receptor-targeted therapeutic approach to counter pathogenic New World mammarenavirus infections.
Nat Commun, 13, 2022
|
|
4RIK
 
 | | Amyloid forming segment, AVVTGVTAV, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 69-77 | | Descriptor: | Alpha-synuclein | | Authors: | Guenther, E.L, Sawaya, M.R, Ivanova, M, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
6CB9
 
 | | Segment AALQSS from the low complexity domain of TDP-43, residues 328-333 | | Descriptor: | AALQSS | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CEW
 
 | | Segment AMMAAA from the low complexity domain of TDP-43, residues 321-326 | | Descriptor: | AMMAAA | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3O7O
 
 | |
4QXX
 
 | |
