5HMX
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 10 | | Descriptor: | 2,2'-(5-(thiophen-2-yl)-1,3-phenylene)diacetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design
J.Med.Chem., 59, 2016
|
|
5HMZ
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 23 | | Descriptor: | 5-(5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl)-4-methoxy-2-methyl-N-(methylsulfonyl)benzamide, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design
J.Med.Chem., 59, 2016
|
|
5HMY
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 15 | | Descriptor: | 2,2'-(5-(5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl)-1,3-phenylene)diacetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design.
J.Med.Chem., 59, 2016
|
|
6OQX
 
 | | Human Liver Receptor Homolog-1 bound to the agonist 5N and a fragment of the Tif2 coregulator | | Descriptor: | (8beta,11alpha,12alpha)-8-(1-phenylethenyl)-1,6:7,14-dicycloprosta-1,3,5,7(14)-tetraen-11-yl sulfamate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
6OR1
 
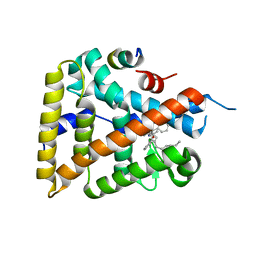 | | Human LRH-1 bound to the agonist 2N and a fragment of the Tif2 coregulator | | Descriptor: | N-[(1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-yl]acetamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
5HMW
 
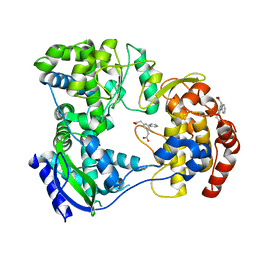 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 5 | | Descriptor: | 2,2'-biphenyl-3,5-diyldiacetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design
J.Med.Chem., 59, 2016
|
|
6OQY
 
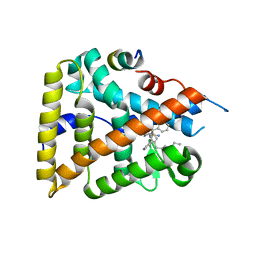 | | Human LRH-1 bound to the agonist 6N and a fragment of the Tif2 coregulator | | Descriptor: | N-[(1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-yl]sulfuric diamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
5HN0
 
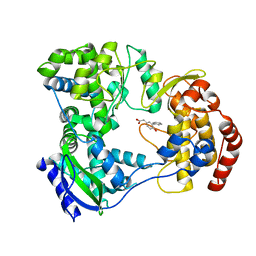 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 4 | | Descriptor: | (6-hydroxybiphenyl-3-yl)acetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design.
J.Med.Chem., 59, 2016
|
|
7BZ9
 
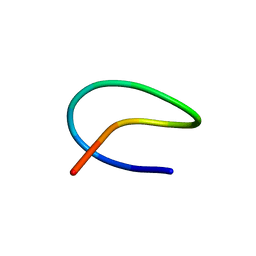 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
3P8Z
 
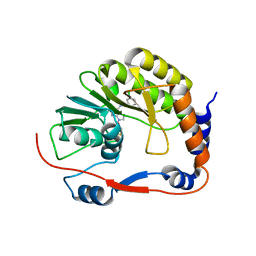 | | Dengue Methyltransferase bound to a SAM-based inhibitor | | Descriptor: | (S)-2-amino-4-(((2S,3S,4R,5R)-5-(6-(3-chlorobenzylamino)-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methylthio)butanoic acid, Non-structural protein 5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Noble, C.G, Yap, L.J, Lescar, J. | | Deposit date: | 2010-10-15 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small molecule inhibitors that selectively block dengue virus methyltransferase
J.Biol.Chem., 286, 2011
|
|
3P97
 
 | |
2NDI
 
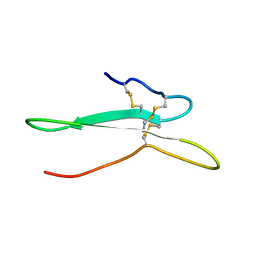 | | Solution structure of the toxin ISTX-I from Ixodes scapularis | | Descriptor: | Putative secreted salivary protein | | Authors: | Hu, K. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A sodium channel inhibitor ISTX-I with a novel structure provides a new hint at the evolutionary link between two toxin folds.
Sci Rep, 6, 2016
|
|
6UH3
 
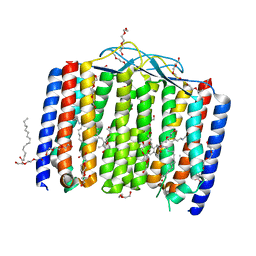 | | Crystal structure of bacterial heliorhodopsin 48C12 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heliorhodopsin, PALMITIC ACID, ... | | Authors: | Lu, Y, Zhou, X.E, Gao, X, Xia, R, Xu, Z, Wang, N, Leng, Y, Melcher, K, Xu, H.E, He, Y. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of heliorhodopsin 48C12.
Cell Res., 30, 2020
|
|
4YVU
 
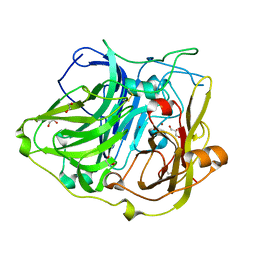 | | Crystal structure of CotA native enzyme in the acid condition, PH5.6 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2015-03-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel binding site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4YVN
 
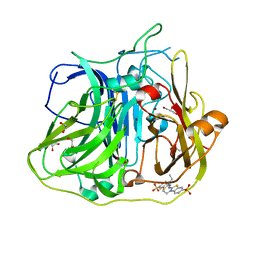 | | Crystal structure of CotA laccase complexed with ABTS at a novel binding site | | Descriptor: | 1,2-ETHANEDIOL, 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, COPPER (II) ION, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2015-03-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel binding site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3E3U
 
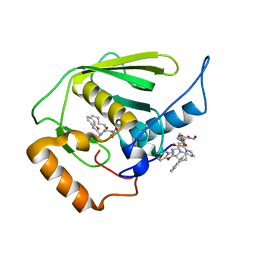 | | Crystal structure of Mycobacterium tuberculosis peptide deformylase in complex with inhibitor | | Descriptor: | N-[(2R)-2-{[(2S)-2-(1,3-benzoxazol-2-yl)pyrrolidin-1-yl]carbonyl}hexyl]-N-hydroxyformamide, NICKEL (II) ION, Peptide deformylase | | Authors: | Meng, W, Xu, M, Pan, S, Koehn, J. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
6A67
 
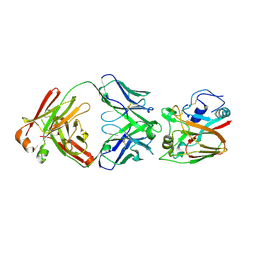 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
5J8V
 
 | |
3NCX
 
 | |
8X3R
 
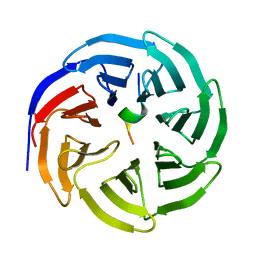 | | Crystal structure of human WDR5 in complex with WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
8X3S
 
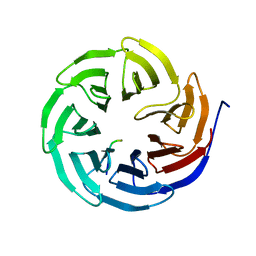 | | Crystal structure of human WDR5 in complex with PTEN | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X, Shang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
3NCW
 
 | |
7T2Q
 
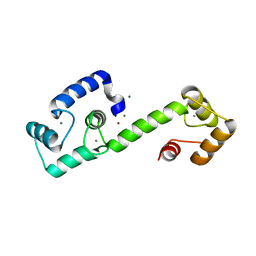 | | PEGylated Calmodulin-1 (K148U) | | Descriptor: | CALCIUM ION, Calmodulin-1, MAGNESIUM ION, ... | | Authors: | Mackay, J.P, Payne, R.J, Patel, K, Dowman, L.J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Site-selective photocatalytic functionalization of peptides and proteins at selenocysteine.
Nat Commun, 13, 2022
|
|
