6KCS
 
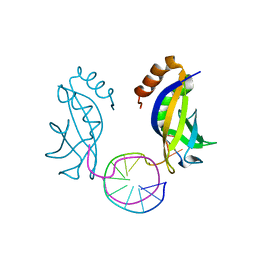 | | Crystal structure of HIRAN domain of HLTF in complex with duplex DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*AP*CP*GP*TP*AP*CP*AP*GP*T)-3'), Helicase-like transcription factor | | Authors: | Hishiki, A, Hashimoto, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of HIRAN domain of human HLTF bound to duplex DNA provides structural basis for DNA unwinding to initiate replication fork regression.
J.Biochem., 167, 2020
|
|
6KX0
 
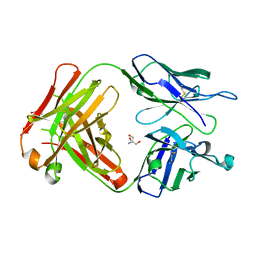 | | Crystal structure of SN-101 mAb non-liganded form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
3ASK
 
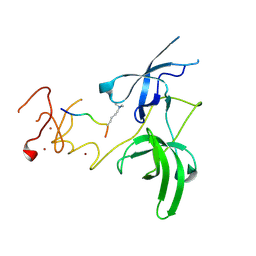 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ASL
 
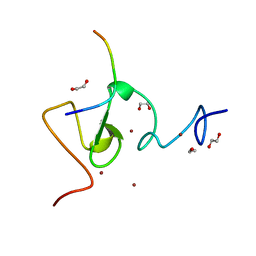 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ... | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3WDN
 
 | |
2EB2
 
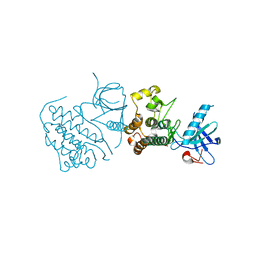 | | Crystal structure of mutated EGFR kinase domain (G719S) | | Descriptor: | Epidermal growth factor receptor | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Senba, K, Yamamoto, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 2012
|
|
2EB3
 
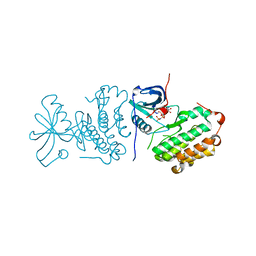 | | Crystal structure of mutated EGFR kinase domain (L858R) in complex with AMPPNP | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Senba, K, Yamamoto, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 2012
|
|
3AIZ
 
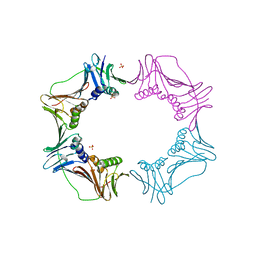 | |
3AIX
 
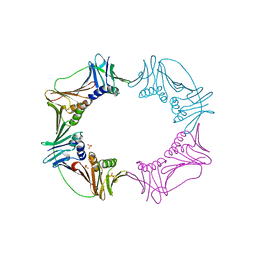 | |
3VWA
 
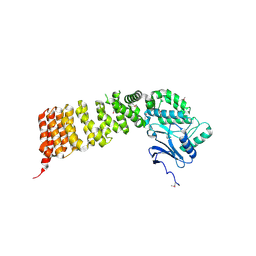 | | Crystal structure of Cex1p | | Descriptor: | Cytoplasmic export protein 1 | | Authors: | Nozawa, K, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Cex1p reveals the mechanism of tRNA trafficking between nucleus and cytoplasm
Nucleic Acids Res., 41, 2013
|
|
3WKL
 
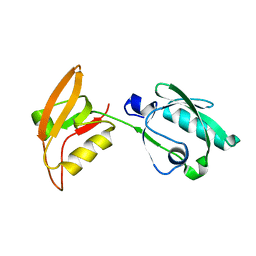 | |
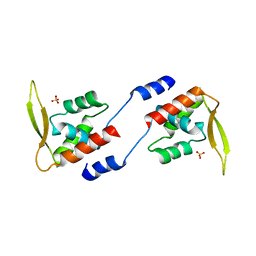
2E1N
 
 | |
3AN2
 
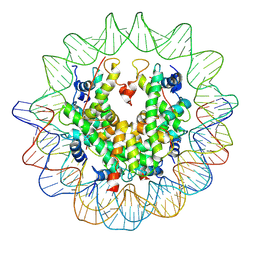 | | The structure of the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tachiwana, H, Kagawa, W, Shiga, T, Saito, K, Osakabe, A, Hayashi-Takanaka, Y, Park, S.-Y, Kimura, H, Kurumizaka, H. | | Deposit date: | 2010-08-27 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the human centromeric nucleosome containing CENP-A
Nature, 476, 2011
|
|
3VJI
 
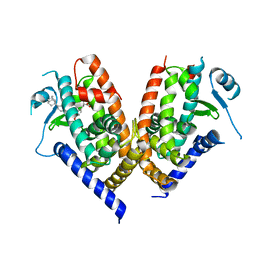 | | Human PPAR gamma ligand binding domain in complex with JKPL53 | | Descriptor: | (2S)-2-{4-butoxy-3-[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]benzoyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
3VJH
 
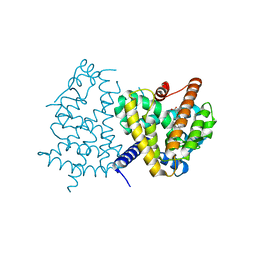 | | Human PPAR GAMMA ligand binding domain in complex with JKPL35 | | Descriptor: | (2S)-2-[4-methoxy-3-({[4-(trifluoromethyl)benzoyl]amino}methyl)benzyl]pentanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
2D9Q
 
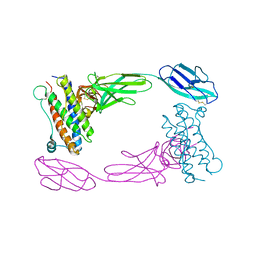 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DLF
 
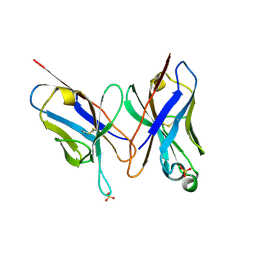 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | Descriptor: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2ZFU
 
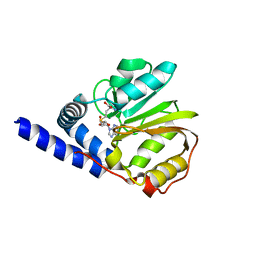 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
3B0T
 
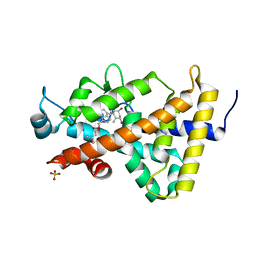 | | Human VDR ligand binding domain in complex with maxacalcitol | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-(3-hydroxy-3-methylbutoxy)-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Hishiki, A, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-06-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human VDR ligand binding domain in complex with maxacalcitol
To be Published
|
|
3B0R
 
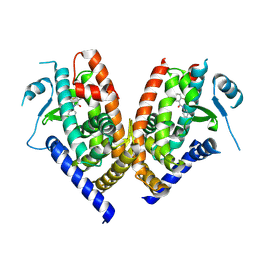 | |
7L38
 
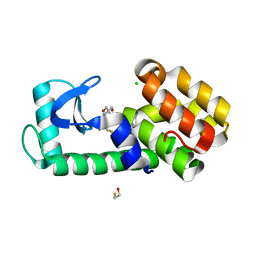 | | T4 Lysozyme L99A - Apo - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L37
 
 | | T4 Lysozyme L99A - Apo - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3J
 
 | | T4 Lysozyme L99A - benzylacetate - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3H
 
 | | T4 Lysozyme L99A - ethylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3A
 
 | | T4 Lysozyme L99A - toluene - cryo | | Descriptor: | Endolysin, TOLUENE | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
