7R08
 
 | | Abortive infection DNA polymerase Abi-P2 | | Descriptor: | Reverse transcriptase | | Authors: | Gapinska, M.A, Figiel, M, Czarnocki Cieciura, M, Nowotny, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
4TNJ
 
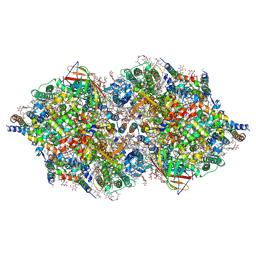 | | RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 4.5 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
7R06
 
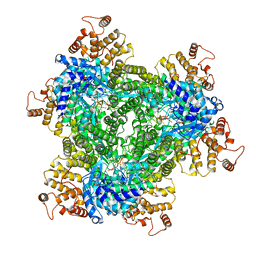 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Figiel, M, Nowotny, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
5G5W
 
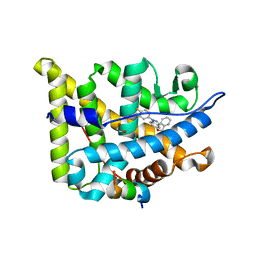 | | Structure guided design and discovery of Indazole ethers as highly potent, non-steroidal Glucocorticoid receptor modulators | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-trifluoro-N-[(1R,2S)-1-{[1-(4-fluorophenyl)-1H-indazol-5-yl]oxy}-1-phenylpropan-2-yl]acetamide, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Hemmerling, M, Edman, K, Lepisto, M, Eriksson, A, Ivanova, S, Dahmen, J, Rehwinkel, H, Berger, M, Hendrickx, R, Dearman, M, Jellesmark-Jensen, T, Wissler, L, Hansson, T. | | Deposit date: | 2016-06-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Indazole Ethers as Novel, Potent, Non-Steroidal Glucocorticoid Receptor Modulators.
Bioorg.Med.Chem.Lett., 26, 2017
|
|
5G2N
 
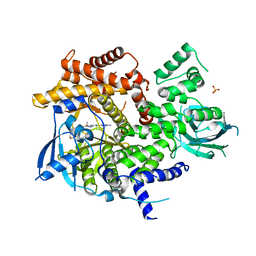 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
7R07
 
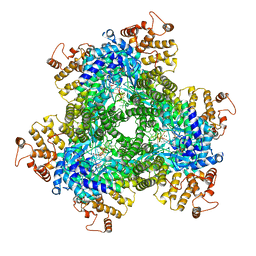 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), MAGNESIUM ION | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7OQV
 
 | |
4TNI
 
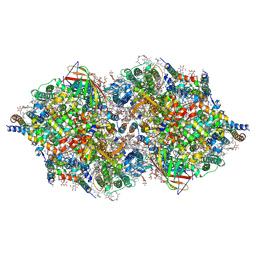 | | RT XFEL structure of Photosystem II 500 ms after the third illumination at 4.6 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNK
 
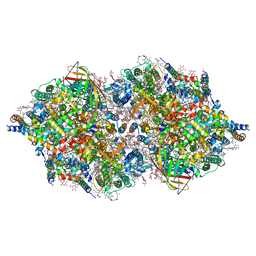 | | RT XFEL structure of Photosystem II 250 microsec after the third illumination at 5.2 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
6XKQ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
7PY2
 
 | | Structure of pathological TDP-43 filaments from ALS with FTLD | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Arseni, D, Hasegawa, H, Murzin, A.G, Kametani, F, Arai, M, Yoshida, M, Falcon, B. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of pathological TDP-43 filaments from ALS with FTLD.
Nature, 601, 2022
|
|
5HNZ
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
8H78
 
 | | Crystal structure of human MMP-2 catalytic domain in complex with inhibitor | | Descriptor: | (2~{R})-2-[[4-[(4-aminocarbonylphenyl)carbonylamino]phenyl]sulfonylamino]-5-[(2~{S},4~{S})-4-azanyl-2-[[(2~{S})-1-[[(2~{S})-1-[(5-azanyl-5-oxidanylidene-pentyl)amino]-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]-methyl-amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamoyl]pyrrolidin-1-yl]-5-oxidanylidene-pentanoic acid, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Takeuchi, T, Mima, M. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of TP0597850: A Selective, Chemically Stable, and Slow Tight-Binding Matrix Metalloproteinase-2 Inhibitor with a Phenylbenzamide-Pentapeptide Hybrid Scaffold.
J.Med.Chem., 66, 2023
|
|
4TTY
 
 | |
5VZY
 
 | | Crystal structure of crenezumab Fab in complex with Abeta | | Descriptor: | Amyloid beta A4 protein, Crenezumab Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Crenezumab Fab light chain,Immunoblobulin light chain | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
5HNW
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2018-07-25 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
7PZC
 
 | | Cryo-EM structure of the NLRP3 decamer bound to the inhibitor CRID3 | | Descriptor: | 1-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Hochheiser, I.V, Pilsl, M, Hagelueken, G, Engel, C, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-26 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.
Nature, 604, 2022
|
|
4TOY
 
 | |
5HNX
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
7PUA
 
 | | Middle assembly intermediate of the Trypanosoma brucei mitoribosomal small subunit | | Descriptor: | 30S Ribosomal protein S17, putative, 30S ribosomal protein S8, ... | | Authors: | Lenarcic, T, Leibundgut, M, Saurer, M, Ramrath, D.J.F, Fluegel, T, Boehringer, D, Ban, N. | | Deposit date: | 2021-09-29 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mitoribosomal small subunit maturation involves formation of initiation-like complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6KQT
 
 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | Descriptor: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
6KQS
 
 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
5WVO
 
 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | Authors: | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | Deposit date: | 2016-12-28 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
7PUB
 
 | | Late assembly intermediate of the Trypanosoma brucei mitoribosomal small subunit | | Descriptor: | 30S Ribosomal protein S17, putative, 30S ribosomal protein S8, ... | | Authors: | Lenarcic, T, Leibundgut, M, Saurer, M, Ramrath, D.J.F, Fluegel, T, Boehringer, D, Ban, N. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mitoribosomal small subunit maturation involves formation of initiation-like complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6XKP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
