8CY4
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-[3-(4-hydroxyphenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 19 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(5-phenylpentyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXZ
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(3-Phenylpropyl)adenosine (Compound 14) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(3-phenylpropyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY0
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MC4756 (Compound 178) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(4-phenylbutyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7WN1
 
 | | Structure of PfNT1(Y190A) in complex with nanobody 48 and inosine | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, INOSINE, nanobody48 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7YDQ
 
 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | Descriptor: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | Authors: | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7C83
 
 | | Crystal structure of an integral membrane steroid 5-alpha-reductase PbSRD5A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ren, R.B, Han, Y.F, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-05-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Nat Commun, 12, 2021
|
|
6E5U
 
 | |
8HJ5
 
 | | Cryo-EM structure of Gq-coupled MRGPRX1 bound with Compound-16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Gan, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2022-11-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of agonist-induced activation of the human itch receptor MRGPRX1.
Plos Biol., 21, 2023
|
|
8JHQ
 
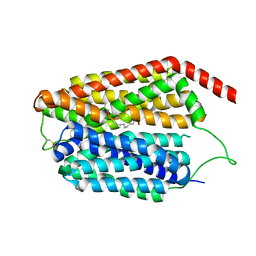 | | Cryo-EM structure of human S1P transporter SPNS2 bound with S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2,GlgA glycogen synthase | | Authors: | Pang, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2023-05-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Spns2-facilitated sphingosine-1-phosphate transport.
Cell Res., 34, 2024
|
|
8JHR
 
 | | Cryo-EM structure of human S1P transporter SPNS2 bound with an inhibitor 16d | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Pang, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2023-05-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Molecular basis of Spns2-facilitated sphingosine-1-phosphate transport.
Cell Res., 34, 2024
|
|
8K2W
 
 | | Structure of CXCR3 complexed with antagonist AMG487 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, N-[(1R)-1-[3-(4-ethoxyphenyl)-4-oxidanylidene-pyrido[2,3-d]pyrimidin-2-yl]ethyl]-N-(pyridin-3-ylmethyl)-2-[4-(trifluoromethyloxy)phenyl]ethanamide, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487.
Cell Discov, 9, 2023
|
|
8K2X
 
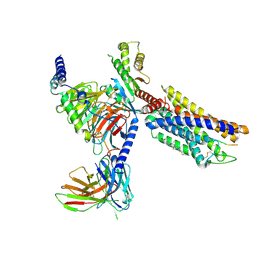 | | CXCR3-DNGi complex activated by CXCL10 | | Descriptor: | C-X-C motif chemokine 10, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487.
Cell Discov, 9, 2023
|
|
6M1I
 
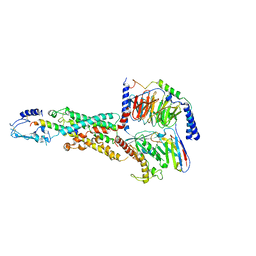 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
6MG1
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta, ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MG3
 
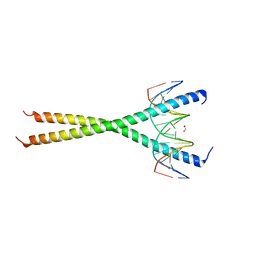 | | V285A Mutant of the C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MG2
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2221 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
8VPI
 
 | |
8VPG
 
 | |
8VPH
 
 | |
7RFK
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor Sinefungin | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFL
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC0946 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFM
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
