7ET7
 
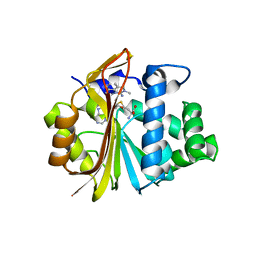 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000028 | | Descriptor: | 10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-12 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
3UA8
 
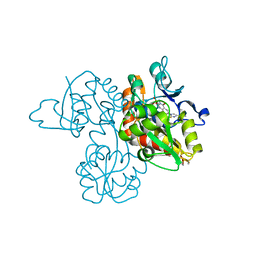 | | Crystal Structure Analysis of a 6-Amino Quinazolinedione Sulfonamide bound to human GluR2 | | Descriptor: | Glutamate receptor 2, N-methyl-1-{3-[(methylsulfonyl)amino]-2,4-dioxo-7-(trifluoromethyl)-1,2,3,4-tetrahydroquinazolin-6-yl}-1H-imidazole-4-carboxamide | | Authors: | Kallen, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Amino quinazolinedione sulfonamides as orally active competitive AMPA receptor antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1QHQ
 
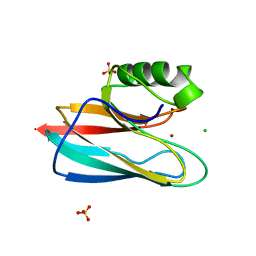 | | AURACYANIN, A BLUE COPPER PROTEIN FROM THE GREEN THERMOPHILIC PHOTOSYNTHETIC BACTERIUM CHLOROFLEXUS AURANTIACUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (AURACYANIN), ... | | Authors: | Bond, C.S, Blankenship, R.E, Freeman, H.C, Guss, J.M, Maher, M, Selvaraj, F, Wilce, M.C.J, Willingham, K. | | Deposit date: | 1999-05-25 | | Release date: | 2001-03-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of auracyanin, a "blue" copper protein from the green thermophilic photosynthetic bacterium Chloroflexus aurantiacus.
J.Mol.Biol., 306, 2001
|
|
1PYA
 
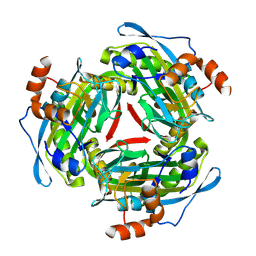 | |
1UOR
 
 | |
5OSW
 
 | | Structure of caprine serum albumin in complex with 3,5-diiodosalicylic acid | | Descriptor: | 2-HYDROXY-3,5-DIIODO-BENZOIC ACID, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, Albumin, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5ORI
 
 | | Structure of caprine serum albumin in orthorhombic crystal system | | Descriptor: | Albumin | | Authors: | Bujacz, A, Talaj, J.A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OTB
 
 | | Structure of caprine serum albumin in P1 space group | | Descriptor: | Albumin, DI(HYDROXYETHYL)ETHER, PROLINE, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5ORF
 
 | | Structure of ovine serum albumin in P1 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, PROLINE, Serum albumin, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2Q7O
 
 | | Structure of human purine nucleoside phosphorylase in complex with L-Immucillin-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2007-06-07 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | L-Enantiomers of transition state analogue inhibitors bound to human purine nucleoside phosphorylase.
J.Am.Chem.Soc., 130, 2008
|
|
1M9B
 
 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with gamma-glutamyl[S-(2-iodobenzyl)cysteinyl]glycine | | Descriptor: | GAMMA-GLUTAMYL[S-(2-IODOBENZYL)CYSTEINYL]GLYCINE, Glutathione S-Transferase 26 kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M99
 
 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with glutathione sulfonic acid | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-Transferase 26kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M9A
 
 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with S-hexylglutathione | | Descriptor: | Glutathione S-Transferase 26 kDa, S-HEXYLGLUTATHIONE | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
4L9K
 
 | | X-ray study of human serum albumin complexed with camptothecin | | Descriptor: | (2S)-2-hydroxy-2-[8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]butanoic acid, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4LB2
 
 | | X-ray study of human serum albumin complexed with idarubicin | | Descriptor: | IDARUBICIN, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4L8U
 
 | | X-ray study of human serum albumin complexed with 9 amino camptothecin | | Descriptor: | (2S)-2-[1-amino-8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]-2-hydroxybutanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-17 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4LA0
 
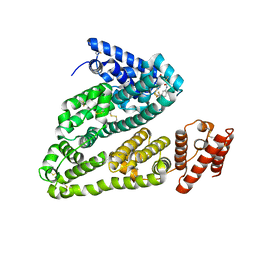 | | X-ray study of human serum albumin complexed with bicalutamide | | Descriptor: | R-BICALUTAMIDE, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4LB9
 
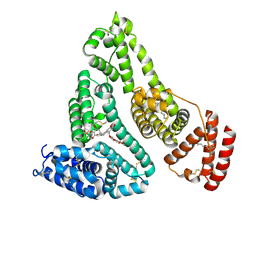 | | X-ray study of human serum albumin complexed with etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol-5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
3ZTF
 
 | | X-ray Structure of the Cyan Fluorescent Protein mTurquoise2 (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
4L9Q
 
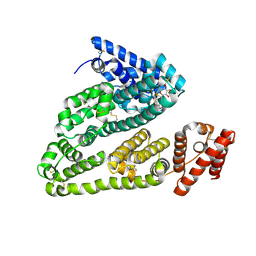 | | X-ray study of human serum albumin complexed with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
1N5U
 
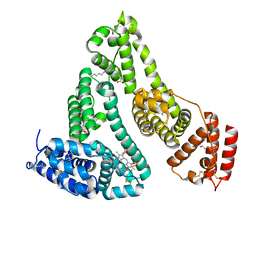 | | X-RAY STUDY OF HUMAN SERUM ALBUMIN COMPLEXED WITH HEME | | Descriptor: | MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SERUM ALBUMIN | | Authors: | Wardell, M, Wang, Z, Ho, J.X, Robert, J, Ruker, F, Ruble, J, Carter, D.C. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Atomic Structure of Human Methemalbumin at 1.9 A
Biochem.Biophys.Res.Commun., 291, 2002
|
|
1PV9
 
 | | Prolidase from Pyrococcus furiosus | | Descriptor: | Xaa-Pro dipeptidase, ZINC ION | | Authors: | Maher, M.J, Ghosh, M, Grunden, A.M, Menon, A.L, Adams, M.W, Freeman, H.C, Guss, J.M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Prolidase from Pyrococcus furiosus.
Biochemistry, 43, 2004
|
|
2EXG
 
 | | Making Protein-Protein Interactions Drugable: Discovery of Low-Molecular-Weight Ligands for the AF6 PDZ Domain | | Descriptor: | (5R)-2-SULFANYL-5-[4-(TRIFLUOROMETHYL)BENZYL]-1,3-THIAZOL-4-ONE, Afadin | | Authors: | Joshi, M, Vargas, C, Boisguerin, P, Krause, G, Schade, M, Oschkinat, H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
3D0T
 
 | |
1H6P
 
 | | Dimeristion domain from human TRF2 | | Descriptor: | MAGNESIUM ION, TELOMERIC REPEAT BINDING FACTOR 2 | | Authors: | Chapman, L, Fairall, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Trfh Dimerization Domain of the Human Telomere Proteins Trf1 and Trf2
Mol.Cell, 8, 2001
|
|
