7NN4
 
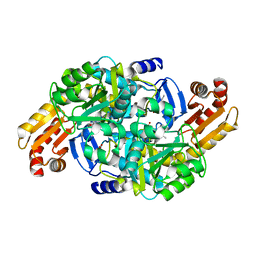 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal 5'-phosphate and 3-hydroxy-2-naphthoic acid. | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NFC
 
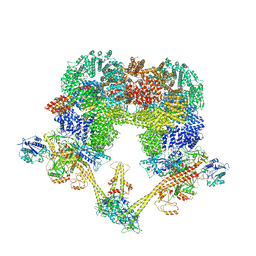 | | Cryo-EM structure of NHEJ super-complex (dimer) | | Descriptor: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NFE
 
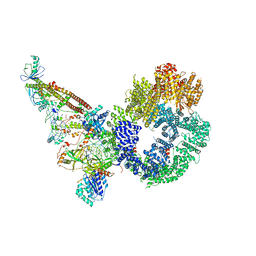 | | Cryo-EM structure of NHEJ super-complex (monomer) | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-06 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7ORZ
 
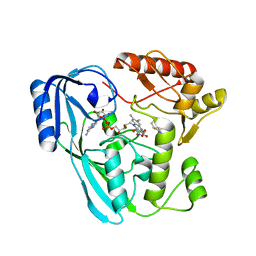 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-06 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OR2
 
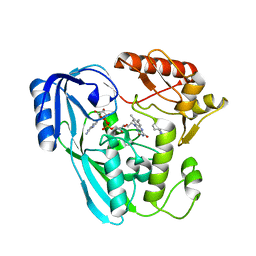 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 4) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OSQ
 
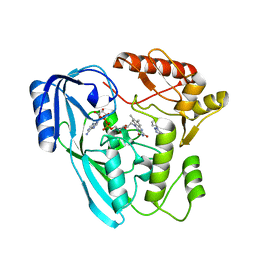 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OTW
 
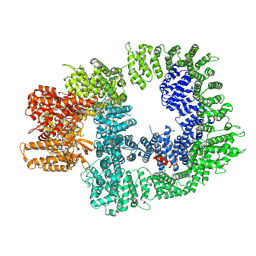 | | DNA-PKcs in complex with AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTV
 
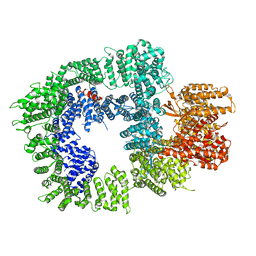 | | DNA-PKcs in complex with wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTM
 
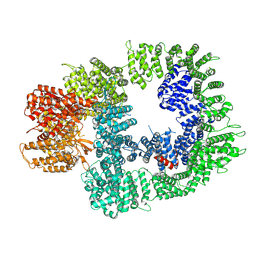 | | Cryo-EM structure of DNA-PKcs in complex with NU7441 | | Descriptor: | 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTY
 
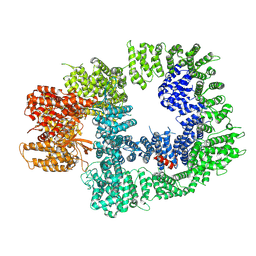 | | DNA-PKcs in complex with M3814 | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTP
 
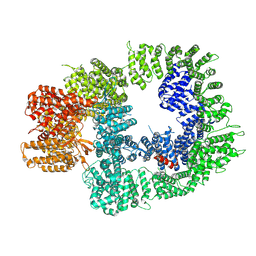 | | DNA-PKcs in complex with ATPgammaS-Mg | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
1EPQ
 
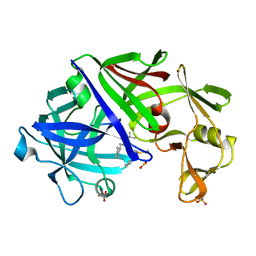 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-133,450 (SOT PHE GLY+SCC GCL) | | Descriptor: | ENDOTHIAPEPSIN, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morp holin-4-ylsulfonyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Dealwis, C, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1SMR
 
 | |
1EPL
 
 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS1, PRO-LEU-GLU-PSA-ARG-LEU | | Authors: | Al-Karadaghi, S, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPP
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-130,693 (MAS PHE LYS+MTF STA MBA) | | Descriptor: | ENDOTHIAPEPSIN, N-(dimethylsulfamoyl)-L-phenylalanyl-N-[(1S,2S)-2-hydroxy-4-{[(2S)-2-methylbutyl]amino}-1-(2-methylpropyl)-4-oxobutyl]-N~6~-(methylcarbamothioyl)-L-lysinamide, SULFATE ION | | Authors: | Wallace, B.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1EPM
 
 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS2, THR-PHE-GLN-ALA-PSA-LEU-ARG-GLU, ... | | Authors: | Crawford, M, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPR
 
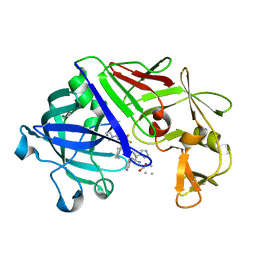 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-135,040 | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Badasso, M, Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPN
 
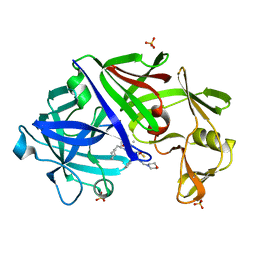 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R,2S)-1-(cyclohexylmethyl)-2-hydroxy-3-(1-methylethoxy)-3-oxopropyl]-S-methyl-L-cysteinamide, SULFATE ION | | Authors: | Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1SGF
 
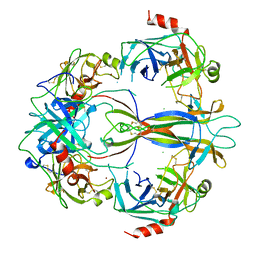 | | CRYSTAL STRUCTURE OF 7S NGF: A COMPLEX OF NERVE GROWTH FACTOR WITH FOUR BINDING PROTEINS (SERINE PROTEINASES) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NERVE GROWTH FACTOR, ... | | Authors: | Bax, B.D.V, Blundell, T.L, Murray-Rust, J, Mcdonald, N.Q. | | Deposit date: | 1997-08-08 | | Release date: | 1998-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of mouse 7S NGF: a complex of nerve growth factor with four binding proteins.
Structure, 5, 1997
|
|
1MPP
 
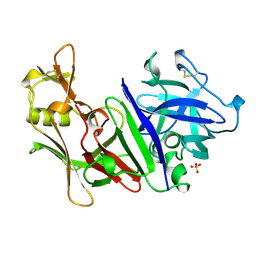 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | Descriptor: | PEPSIN, SULFATE ION | | Authors: | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
1N0W
 
 | | Crystal structure of a RAD51-BRCA2 BRC repeat complex | | Descriptor: | 1,2-ETHANEDIOL, ARTIFICIAL GLY-SER-MSE-GLY PEPTIDE, Breast cancer type 2 susceptibility protein, ... | | Authors: | Pellegrini, L, Yu, D.S, Lo, T, Anand, S, Lee, M, Blundell, T.L, Venkitaraman, A.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into DNA recombination from the structure of a RAD51-BRCA2 complex
Nature, 420, 2002
|
|
1NK1
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
1M3U
 
 | | Crystal Structure of Ketopantoate Hydroxymethyltransferase complexed the Product Ketopantoate | | Descriptor: | 3-methyl-2-oxobutanoate hydroxymethyltransferase, KETOPANTOATE, MAGNESIUM ION | | Authors: | von Delft, F, Inoue, T, Saldanha, S.A, Ottenhof, H.H, Dhanaraj, V, Witty, M, Abell, C, Smith, A.G, Blundell, T.L. | | Deposit date: | 2002-06-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E. coli Ketopantoate Hydroxymethyl Transferase Complexed with Ketopantoate and Mg(2+), Solved by Locating 160 Selenomethionine Sites.
Structure, 11, 2003
|
|
1PT1
 
 | | Unprocessed Pyruvoyl Dependent Aspartate Decarboxylase with Histidine 11 Mutated to Alanine | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-22 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1SAC
 
 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
