7XR3
 
 | |
4JZR
 
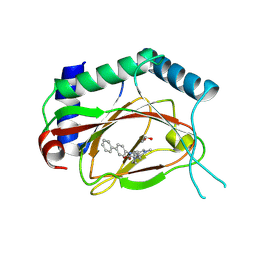 | | Structure of Prolyl Hydroxylase Domain-containing Protein (PHD) with Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-(biphenyl-4-yl)-8-[(1-methyl-1H-imidazol-2-yl)methyl]-2,8-diazaspiro[4.5]decan-1-one, Egl nine homolog 1, ... | | Authors: | Ma, Y, Yang, L. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel complex crystal structure of prolyl hydroxylase domain-containing protein 2 (PHD2): 2,8-Diazaspiro[4.5]decan-1-ones as potent, orally bioavailable PHD2 inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
6DNE
 
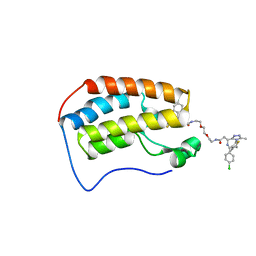 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS660 | | Descriptor: | Bromodomain-containing protein 4, N,N'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4ZHV
 
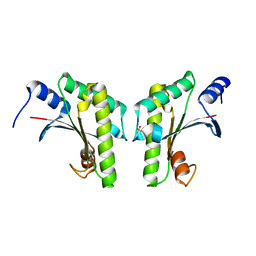 | | Crystal structure of a bacterial signalling protein | | Descriptor: | SULFATE ION, YfiB | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
8S9H
 
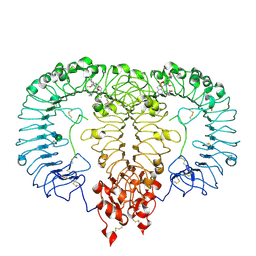 | | Crystal structure of monkey TLR7 ectodomain with compound 14 | | Descriptor: | (3S)-3-{[5-amino-1-({3-methoxy-5-[1-(oxan-4-yl)piperidin-4-yl]pyridin-2-yl}methyl)-1H-pyrazolo[4,3-d]pyrimidin-7-yl]amino}hexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Critton, D.A. | | Deposit date: | 2023-03-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Identification and Optimization of Small Molecule Pyrazolopyrimidine TLR7 Agonists for Applications in Immuno-oncology.
Acs Med.Chem.Lett., 15, 2024
|
|
6DJC
 
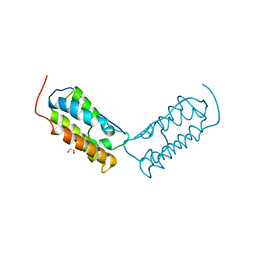 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2MJV
 
 | |
4WD9
 
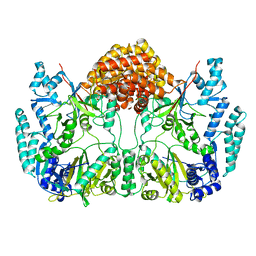 | |
4WNT
 
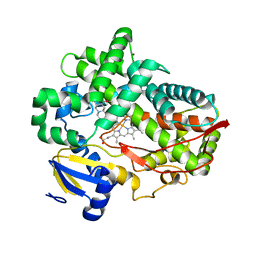 | | Human Cytochrome P450 2D6 Ajmalicine Complex | | Descriptor: | Ajmalicine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
5L1I
 
 | |
5L01
 
 | | Tryptophan 5-hydroxylase in complex with inhibitor (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid | | Descriptor: | (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5L1L
 
 | | PostInsertion complex of Human DNA Polymerase Eta bypassing an O6-Methyl-2'-deoxyguanosine : dT site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*(6OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2'-deoxyguanosine by Human DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
4ZV8
 
 | | Structure of CYP2B6 (Y226H/K262R) with additional mutation Y244W in complex with alpha-Pinene | | Descriptor: | (+)-alpha-Pinene, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Liu, J, Shah, M.B, Stout, C.D, Halpert, J.R. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Coumarin Derivatives as Substrate Probes of Mammalian Cytochromes P450 2B4 and 2B6: Assessing the Importance of 7-Alkoxy Chain Length, Halogen Substitution, and Non-Active Site Mutations.
Biochemistry, 55, 2016
|
|
5L1K
 
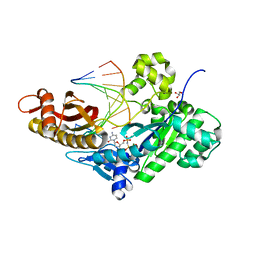 | | PostInsertion complex of Human DNA Polymerase Eta bypassing an O6-Methyl-2'-deoxyguanosine : dC site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*GP*(6OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2'-deoxyguanosine by Human DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5L1J
 
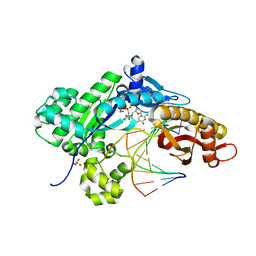 | |
5KQE
 
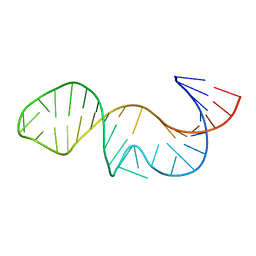 | |
6DUV
 
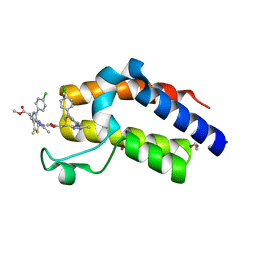 | | Crystal structure of the second bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Babault, N, Zhou, M.M. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Bound Water Molecules in Differential Recognition of Di-Acetylated Histone Peptides by the BRD4 Bromodomains
To be published
|
|
5I9H
 
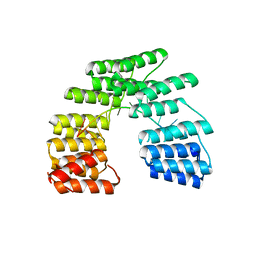 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9F
 
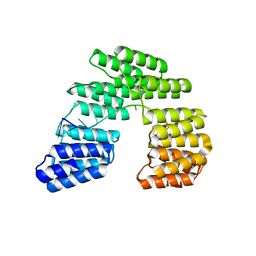 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U10 in complex with its target RNA U10 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U10 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, Y. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9D
 
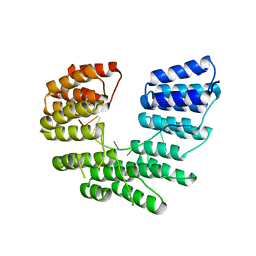 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8A2 in complex with its target RNA U8A2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*G*UP*UP*UP*UP*AP*AP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8A2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
