5CRZ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-4-fluorobenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5D0C
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-08-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CTL
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)benzenesulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
7Y8R
 
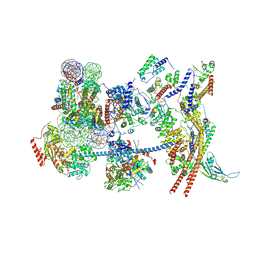 | | The nucleosome-bound human PBAF complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2, ... | | Authors: | Wang, L, Yu, J, Yu, Z, Wang, Q, He, S, Xu, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of nucleosome-bound human PBAF complex.
Nat Commun, 13, 2022
|
|
5JO7
 
 | | Henbane premnaspirodiene synthase (HPS), also known as Henbane vetispiradiene synthase (HVS) from Hyoscyamus muticus | | Descriptor: | Vetispiradiene synthase 1 | | Authors: | Koo, H.J, Xu, Y, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional study of terpene synthase from 512 mutant library of henbane premnaspirodiene synthase reveals protein residue interactions
To Be Published
|
|
1RJI
 
 | | Solution Structure of BmKX, a novel potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | potassium channel toxin KX | | Authors: | Cai, Z, Wu, J, Xu, Y, Wang, C.-G, Chi, C.-W, Shi, Y. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A novel short-chain peptide BmKX from the Chinese scorpion Buthus martensi karsch, sequencing, gene cloning and structure determination
Toxicon, 45, 2005
|
|
1DLE
 
 | | FACTOR B SERINE PROTEASE DOMAIN | | Descriptor: | COMPLEMENT FACTOR B | | Authors: | Jing, H, Xu, Y, Carson, M, Moore, D, Macon, K.J, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 1999-12-09 | | Release date: | 2000-12-13 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New structural motifs on the chymotrypsin fold and their potential roles in complement factor B.
EMBO J., 19, 2000
|
|
4PY4
 
 | |
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
3P0U
 
 | | Crystal Structure of the ligand binding domain of human testicular receptor 4 | | Descriptor: | Nuclear receptor subfamily 2 group C member 2 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Xu, Y, Chan, C.-W, Kruse, S.W, Reynolds, R, Engel, J.D, Xu, H.E. | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Orphan Nuclear Receptor TR4 Is a Vitamin A-activated Nuclear Receptor.
J.Biol.Chem., 286, 2011
|
|
2YKR
 
 | | 30S ribosomal subunit with RsgA bound in the presence of GMPPNP | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Guo, Q, Yuan, Y, Xu, Y, Feng, B, Liu, L, Chen, K, Lei, J, Gao, N. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural Basis for the Function of a Small Gtpase Rsga on the 30S Ribosomal Subunit Maturation Revealed by Cryoelectron Microscopy.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8QN0
 
 | | Soluble epoxide hydrolase in complex with RK3 | | Descriptor: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-5-(4-methylphenyl)sulfonyl-1,3,3~{a},4,6,6~{a}-hexahydropyrrolo[3,4-c]pyrrole-2-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-25 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
8QMZ
 
 | | Soluble epoxide hydrolase in complex with RK4 | | Descriptor: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-2-(4-methylphenyl)sulfonyl-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-5-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-25 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
3L1N
 
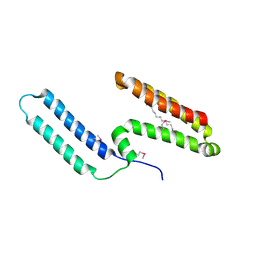 | | Crystal structure of Mp1p ligand binding domain 2 complexd with palmitic acid | | Descriptor: | Cell wall antigen, PALMITIC ACID | | Authors: | Liao, S, Tung, E.T, Zheng, W, Chong, K, Xu, Y, Bartlam, M, Rao, Z, Yuen, K.Y. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the Mp1p ligand binding domain 2 reveals its function as a fatty acid-binding protein.
J.Biol.Chem., 285, 2010
|
|
5H1B
 
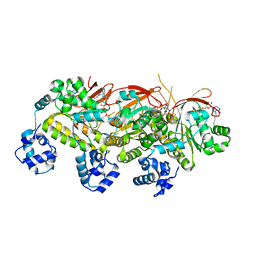 | | Human RAD51 presynaptic complex | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, MAGNESIUM ION, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
8HK5
 
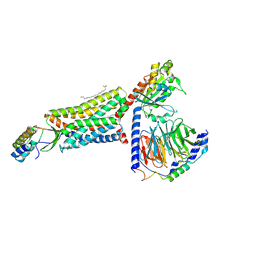 | | C5aR1-Gi-C5a protein complex | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Complement C5, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK3
 
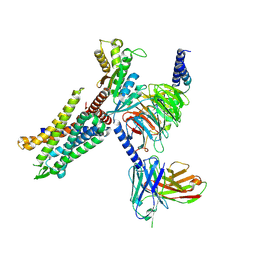 | | C3aR-Gi-apo protein complex | | Descriptor: | C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK2
 
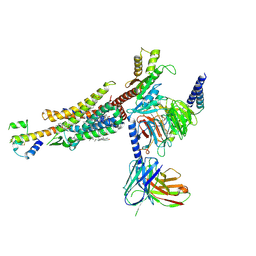 | | C3aR-Gi-C3a protein complex | | Descriptor: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
5H1C
 
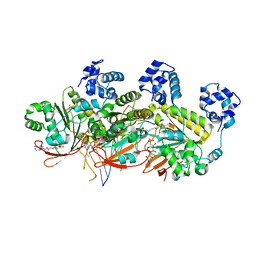 | | Human RAD51 post-synaptic complexes | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
4RPP
 
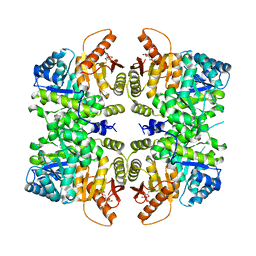 | | crystal structure of PKM2-K422R mutant bound with FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
8IBU
 
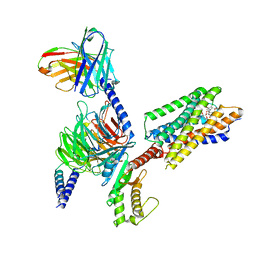 | | Cryo-EM structure of the erythromycin-bound motilin receptor-Gq protein complex | | Descriptor: | ERYTHROMYCIN A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | You, C, Jiang, Y, Xu, H.E, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
8IBV
 
 | | Cryo-EM structure of the motilin-bound motilin receptor-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Jiang, Y, Xu, H.E, You, C, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
1HIY
 
 | | Binding of nucleotides to NDP kinase | | Descriptor: | 3'-DEOXY 3'-AMINO ADENOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Cervoni, L, Lascu, I, Xu, Y, Gonin, P, Morr, M, Merouani, M, Janin, J, Giartoso, A. | | Deposit date: | 2001-01-05 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of Nucleotides to Nucleoside Diphosphate Kinase: A Calorimetric Study.
Biochemistry, 40, 2001
|
|
1KSB
 
 | | Relationship of Solution and Protein-Bound Structures of DNA Duplexes with the Major Intrastrand Cross-Link Lesions Formed on Cisplatin Binding to DNA | | Descriptor: | 5'-D(*AP*GP*GP*CP*CP*GP*GP*AP*G)-3', 5'-D(*CP*TP*CP*CP*GP*GP*CP*CP*T)-3', Cisplatin | | Authors: | Marzilli, L.G, Saad, J.S, Kuklenyik, Z, Keating, K.A, Xu, Y. | | Deposit date: | 2002-01-11 | | Release date: | 2002-01-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Relationship of solution and protein-bound structures of DNA duplexes with the major intrastrand cross-link lesions formed on cisplatin binding to DNA.
J.Am.Chem.Soc., 123, 2001
|
|
8GUD
 
 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E545K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
