9LO7
 
 | | The crystal structure of PDE4D with 2317b | | Descriptor: | 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huag, Y.-Y, Wu, D, Luo, H.-B. | | Deposit date: | 2025-01-22 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.20006251 Å) | | Cite: | Structure-Based Optimization of Moracin M as Potent and Selective PDE4 Inhibitors with Antipsoriasis Effects.
J.Med.Chem., 68, 2025
|
|
5KCJ
 
 | | Structure of the human GluN1/GluN2A LBD in complex with GNE6901 | | Descriptor: | 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
5L1Z
 
 | | TAR complex with HIV-1 Tat-AFF4-P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze-Gahmen, U, Hurley, J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.
Elife, 5, 2016
|
|
3J98
 
 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J97
 
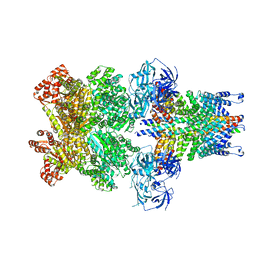 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
9LSM
 
 | | The crystal structure of PDE5A with L9 | | Descriptor: | 1-[(13~{S},15~{R})-4-bromanyl-15-(3-chloranyl-4-methoxy-phenyl)-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaen-12-yl]ethanone, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
9LSL
 
 | | The crystal structure of PDE5A with L1 | | Descriptor: | (13~{S},15~{R})-15-(3-chloranyl-4-methoxy-phenyl)-12-ethanoyl-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaene-4-carbonitrile, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
6M6X
 
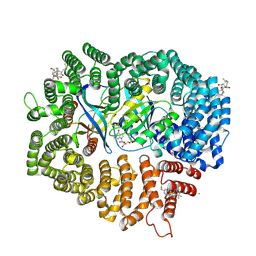 | | Oridonin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
6M60
 
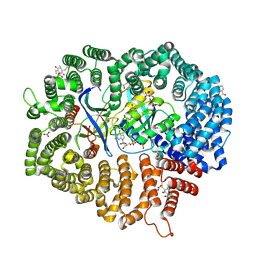 | | Plumbagin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (2~{R})-2-methyl-5-oxidanyl-2,3-dihydronaphthalene-1,4-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
6MEV
 
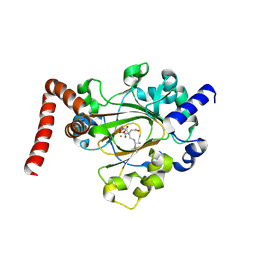 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
9L5T
 
 | | Cryo-EM structure of the thermophile spliceosome (state B*Q2) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5S
 
 | | Cryo-EM structure of the thermophile spliceosome (state B*Q1) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5R
 
 | | Cryo-EM structure of the thermophile spliceosome (state ILS) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Cell cycle control protein, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
3J99
 
 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J96
 
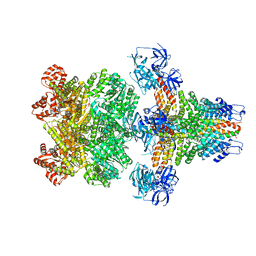 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J95
 
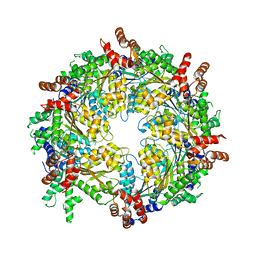 | | Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Vesicle-fusing ATPase | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3J94
 
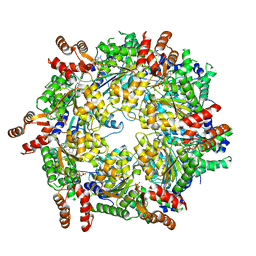 | | Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vesicle-fusing ATPase | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
2GP4
 
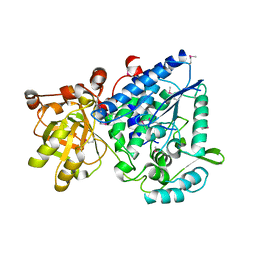 | |
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
8ZUB
 
 | | The Crystal structure of mol075 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-pentyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2024-06-08 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of pyrimidone derivatives as nonpeptidic and noncovalent 3-chymotrypsin-like protease (3CL pro ) inhibitors with anti-coronavirus activities.
Bioorg.Chem., 154, 2025
|
|
8ZUC
 
 | | The Crystal structure of mol080 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[[6-chloranyl-2-(3-methylbutyl)indazol-5-yl]amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2024-06-08 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of pyrimidone derivatives as nonpeptidic and noncovalent 3-chymotrypsin-like protease (3CL pro ) inhibitors with anti-coronavirus activities.
Bioorg.Chem., 154, 2025
|
|
8ZT9
 
 | | The Crystal structure of mol066 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-propan-2-yl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione, GLYCEROL | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2024-06-06 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of pyrimidone derivatives as nonpeptidic and noncovalent 3-chymotrypsin-like protease (3CL pro ) inhibitors with anti-coronavirus activities.
Bioorg.Chem., 154, 2025
|
|
7FIN
 
 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, GAMMA-L-GLUTAMIC ACID, Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-07-31 | | Release date: | 2022-02-23 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
