8TDI
 
 | | Structure of P2B11 Glucuronide-3-dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, P2B11 Glucuronide-3-dehydrogenase, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-03 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
7PX1
 
 | | Conotoxin from Conus mucronatus | | Descriptor: | CADMIUM ION, Conus mucronatus, IODIDE ION | | Authors: | Mueller, E, Hackney, C.M, Ellgaard, L, Morth, J.P. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A previously unrecognized superfamily of macro-conotoxins includes an inhibitor of the sensory neuron calcium channel Cav2.3.
Plos Biol., 21, 2023
|
|
7PX2
 
 | | Conotoxin Mu8.1 from Conus mucronatus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Conotoxin Mu8.1 | | Authors: | Mueller, E, Hackney, C, Ellgaard, L, Morth, J.P. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A previously unrecognized superfamily of macro-conotoxins includes an inhibitor of the sensory neuron calcium channel Cav2.3.
Plos Biol., 21, 2023
|
|
5V7R
 
 | |
5V7U
 
 | |
7BGC
 
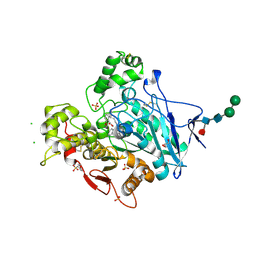 | | human butyrylcholinesterase in complex with a tacrine-methylanacardate hybrid inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Salerno, A, Bolognesi, M.L. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sustainable Drug Discovery of Multi-Target-Directed Ligands for Alzheimer's Disease.
J.Med.Chem., 64, 2021
|
|
4Y8D
 
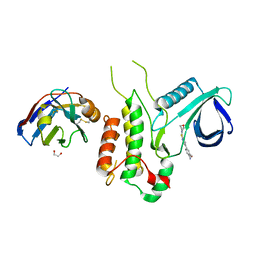 | | Crystal structure of Cyclin-G associated kinase (GAK) complexed with selective 12i inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-[3-(morpholin-4-yl)[1,2]thiazolo[4,3-b]pyridin-6-yl]aniline, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Heroven, C, Nowak, R, De Jonghe, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
J.Med.Chem., 58, 2015
|
|
5HKG
 
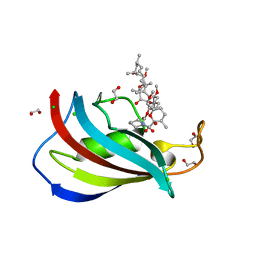 | | Total chemical synthesis, refolding and crystallographic structure of a fully active immunophilin: calstabin 2 (FKBP12.6). | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Sirigu, S, Huet, T, Bacchi, M, Jullian, M, Fould, B, Ferry, G, Vuillard, L, Chavas, L, Puget, K, Nosjean, O, Boutin, J.A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Total chemical synthesis, refolding, and crystallographic structure of fully active immunophilin calstabin 2 (FKBP12.6).
Protein Sci., 25, 2016
|
|
6ZK1
 
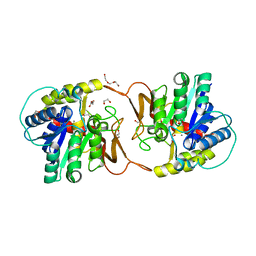 | | Plant nucleoside hydrolase - ZmNRh2b enzyme | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK2
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK3
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with ribose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6X58
 
 | | MPER-Fluc-Ec2 bound to 10E8v4 antibody | | Descriptor: | 10E8v4 Fab Heavy Chain, 10E8v4 Fab Light Chain, gp41 MPER peptide,Putative fluoride ion transporter CrcB | | Authors: | McIlwain, B.C, Stockbridge, R.B. | | Deposit date: | 2020-05-25 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | N-terminal Transmembrane-Helix Epitope Tag for X-ray Crystallography and Electron Microscopy of Small Membrane Proteins.
J.Mol.Biol., 433, 2021
|
|
8TCS
 
 | | Structure of trehalose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, Xylose isomerase-like TIM barrel domain-containing protein, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TDH
 
 | |
8QTL
 
 | | Aplysia californica acetylcholine-binding protein in complex with Spiroimine (-)-4 S | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Soluble acetylcholine receptor, ... | | Authors: | Sulzenbacher, G, Bourne, Y, Marchot, P. | | Deposit date: | 2023-10-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cyclic Imine Core Common to the Marine Macrocyclic Toxins Is Sufficient to Dictate Nicotinic Acetylcholine Receptor Antagonism.
Mar Drugs, 22, 2024
|
|
5XWR
 
 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8V31
 
 | |
8QX2
 
 | |
6ZK4
 
 | | Plant nucleoside hydrolase - ZmNRh2b with a bound adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK5
 
 | | Plant nucleoside hydrolase - ZmNRh3 enzyme in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
8Q1M
 
 | | Aplysia californica acetylcholine-binding protein in complex with Spiroimine (+)-4 R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Soluble acetylcholine receptor, ... | | Authors: | Sulzenbacher, G, Bourne, Y, Marchot, P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Cyclic Imine Core Common to the Marine Macrocyclic Toxins Is Sufficient to Dictate Nicotinic Acetylcholine Receptor Antagonism.
Mar Drugs, 22, 2024
|
|
7Z8Q
 
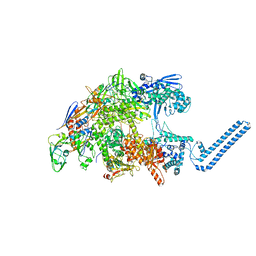 | | Cryo-EM structure of Mycobacterium tuberculosis RNA polymerase core | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Brodolin, K. | | Deposit date: | 2022-03-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization
Nat Commun, 14, 2023
|
|
4PZW
 
 | | Synthesis, Characterization and PK/PD Studies of a Series of Spirocyclic Pyranochromene BACE1 Inhibitors | | Descriptor: | (4R,4a'S,10a'S)-7'-(5-chloropyridin-3-yl)-3',4',4a',10a'-tetrahydro-1'H-spiro[1,3-oxazole-4,5'-pyrano[3,4-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-05-14 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, characterization, and PK/PD studies of a series of spirocyclic pyranochromene BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3U9Q
 
 | |
8SZ8
 
 | |
