7A6U
 
 | | Cryo-EM structure of the cytoplasmic domain of human TRPC6 | | Descriptor: | Short transient receptor potential channel 6, UNKNOWN ATOM OR ION | | Authors: | Grieben, M, Pike, A.C.W, Wang, D, Mukhopadhyay, S.M.M, Chalk, R, Marsden, B.D, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structure of the cytoplasmic domain of human TRPC6
TO BE PUBLISHED
|
|
6RU7
 
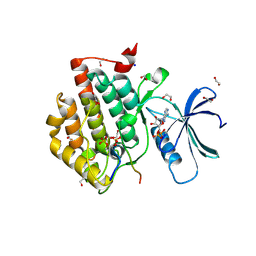 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with double phosphorylated p63 PAD2P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
6P9G
 
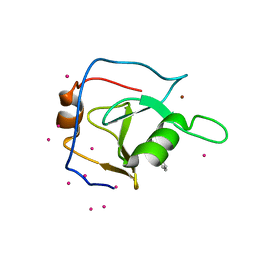 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 2-(4-oxoquinazolin-3(4H)-yl)propanoic acid | | Descriptor: | (2R)-2-(4-oxoquinazolin-3(4H)-yl)propanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Tempel, W, Mann, M.K, Harding, R.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
6SFI
 
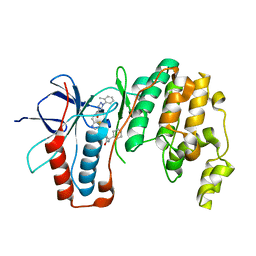 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SFJ
 
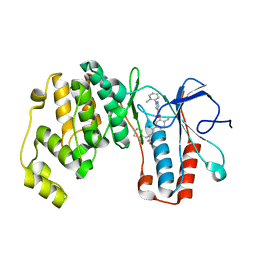 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SE4
 
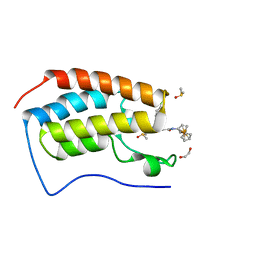 | | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor | | Descriptor: | (+)-JD1, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Hassell-Hart, S, Picaud, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Spencer, J, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor
To Be Published
|
|
6Q3Z
 
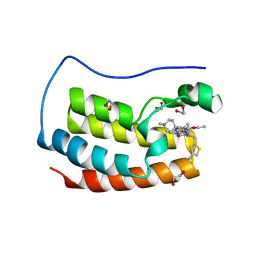 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16k | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-[(4-methylthiophen-2-yl)methyl]-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
4EZA
 
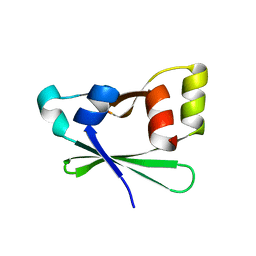 | | Crystal structure of the atypical phosphoinositide (aPI) binding domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2 | | Authors: | Van Aalten, D.M.F, Dixon, M.J, Gray, A, Schenning, M, Agacan, M, Leslie, N.R, Downes, C.P, Batty, I.H, Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold.
J.Biol.Chem., 287, 2012
|
|
6T6D
 
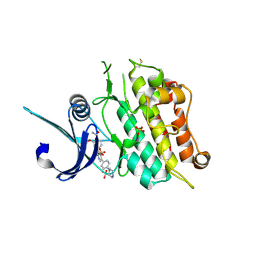 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2149 | | Descriptor: | 2-methoxy-4-[4-methyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]benzamide, Activin receptor type I, SULFATE ION | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
6Q3Y
 
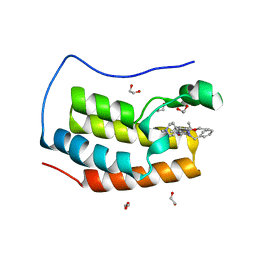 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16i | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-(phenylmethyl)-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
6Q8K
 
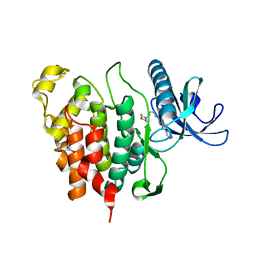 | | CLK1 with bound pyridoquinazoline | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, ~{N}2-(3-morpholin-4-ylpropyl)pyrido[3,4-g]quinazoline-2,10-diamine | | Authors: | Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Joesselin, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
6TCX
 
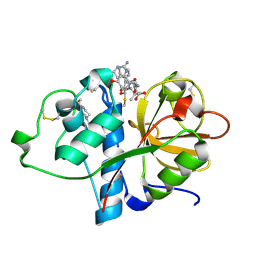 | | Papain bound to a natural cysteine protease inhibitor from Streptomyces mobaraensis | | Descriptor: | (2~{R})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-3-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-(4-hydroxyphenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Papain | | Authors: | Kraemer, A, Juettner, N.E, Fuchsbauer, H.-L, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding the Papain Inhibitor from Streptomyces mobaraensis as Being Hydroxylated Chymostatin Derivatives: Purification, Structure Analysis, and Putative Biosynthetic Pathway.
J.Nat.Prod., 83, 2020
|
|
6TT5
 
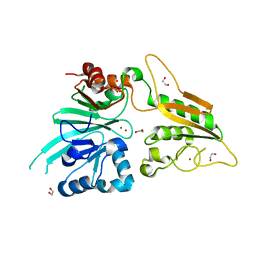 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6QB5
 
 | | Crystal structure of the N-terminal region of human cohesin subunit STAG1 | | Descriptor: | Cohesin subunit SA-1, SODIUM ION | | Authors: | Newman, J.A, Katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-12-20 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
6TSG
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG) in complex with TETRAC | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Gellrich, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | l-Thyroxin and the Nonclassical Thyroid Hormone TETRAC Are Potent Activators of PPAR gamma.
J.Med.Chem., 63, 2020
|
|
4DO0
 
 | | Crystal Structure of human PHF8 in complex with Daminozide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DAMINOZIDE, ... | | Authors: | Krojer, T, Daniel, M, Ng, S.S, Walport, L.J, Chowdhury, R, Arrowsmith, C.H, Edwards, A, Bountra, C, Kawamura, A, Muller-Knapp, S, McDonough, M.A, von Delft, F, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: |
|
|
6QXK
 
 | | Human PIM1 bound to OX0999 | | Descriptor: | 2-[(1-methylpiperidin-4-yl)methylamino]-5-[[2-[4-(trifluoromethyloxy)phenyl]-1,3-thiazol-4-yl]methyl]-1,3-thiazol-4-one, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Alexander, L.T, Elkins, J.M, Russell, A, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PIM1 bound to OX0999
To Be Published
|
|
6TXS
 
 | | The structure of the FERM domain and helical linker of human moesin bound to a CD44 peptide | | Descriptor: | CD44 antigen, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
6SZM
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2009 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-methyl-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl]piperazine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
6T1L
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 3 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(diethylaminomethyl)phenyl]methyl]-4-pyrimidin-2-yl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T6B
 
 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
6R65
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
4FLP
 
 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain testis-specific protein, POTASSIUM ION | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Canning, P, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bradner, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-15 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Small-Molecule Inhibition of BRDT for Male Contraception.
Cell(Cambridge,Mass.), 150, 2012
|
|
4FK7
 
 | | Crystal structure of Certhrax catalytic domain | | Descriptor: | CHLORIDE ION, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-12 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
