4RC7
 
 | |
7W9P
 
 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV pi helix conformer) | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9M
 
 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (S6IV pi helix conformer) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9L
 
 | | Cryo-EM structure of human Nav1.7(E406K)-beta1-beta2 complex | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9T
 
 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV alpha helix conformer) | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9K
 
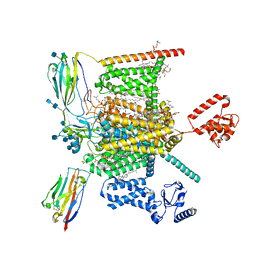 | | Cryo-EM structure of human Nav1.7-beta1-beta2 complex at 2.2 angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7Y89
 
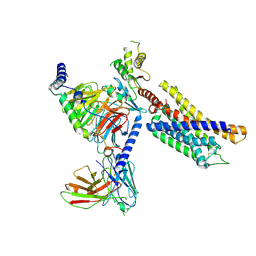 | | Structure of the GPR17-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Ye, F, Chen, G. | | Deposit date: | 2022-06-23 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure of G-protein-coupled receptor GPR17 in complex with inhibitory G protein.
MedComm (2020), 3, 2022
|
|
8ZEE
 
 | | Cryo-EM structure of an intermediate-state PSII-PRF2' complex during the process of photosystem II repair | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, A, Liu, Z. | | Deposit date: | 2024-05-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for an early stage of the photosystem II repair cycle in Chlamydomonas reinhardtii.
Nat Commun, 15, 2024
|
|
6U09
 
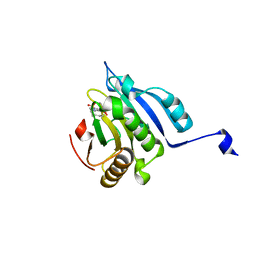 | |
6U06
 
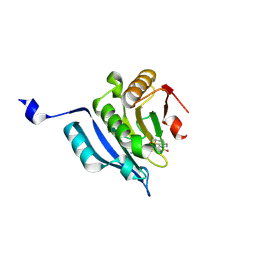 | |
3QQ2
 
 | |
7KS9
 
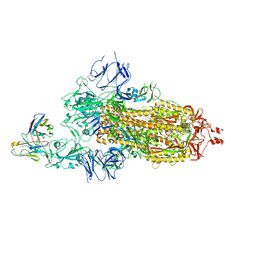 | |
6IW2
 
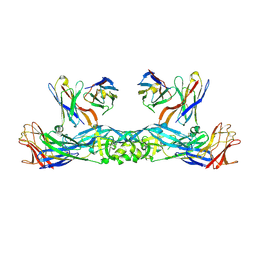 | | Crystal structure of 5A ScFv in complex with YFV-17D sE in prefusion state | | Descriptor: | Envelope protein E, Heavy chain of monoclonal antibody 5A, Light chain of monoclonal antibody 5A | | Authors: | Lu, X.S, Xiao, H.X, Li, S.H, Pang, X.F. | | Deposit date: | 2018-12-04 | | Release date: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Cell Rep, 26, 2019
|
|
3J1E
 
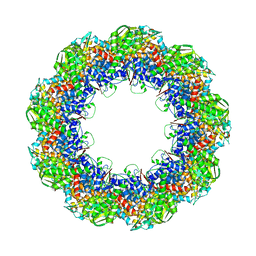 | | Cryo-EM structure of 9-fold symmetric rATcpn-beta in apo state | | Descriptor: | Chaperonin beta subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1B
 
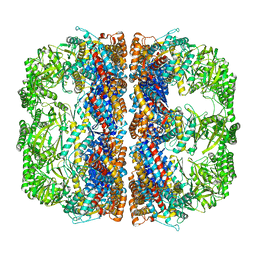 | | Cryo-EM structure of 8-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1C
 
 | | Cryo-EM structure of 9-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
5H3D
 
 | | Helical structure of membrane tubules decorated by ACAP1 (BARPH doamin) protein by cryo-electron microscopy and MD simulation | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Chan, C, Pang, X.Y, Zhang, Y, Sun, F, Fan, J. | | Deposit date: | 2016-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Plos Comput.Biol., 15, 2019
|
|
3J1F
 
 | | Cryo-EM structure of 9-fold symmetric rATcpn-beta in ATP-binding state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin beta subunit, MAGNESIUM ION | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3SFD
 
 | | crystal structure of porcine mitochondrial respiratory complex II bound with oxaloacetate and pentachlorophenol | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhou, Q.J, Zhai, Y.J, Liu, M, Sun, F. | | Deposit date: | 2011-06-13 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex II via a water molecule mediated binding feature.
Protein Cell, 2, 2011
|
|
3SFE
 
 | | crystal structure of porcine mitochondrial respiratory complex II bound with oxaloacetate and thiabendazole | | Descriptor: | 2-(1,3-THIAZOL-4-YL)-1H-BENZIMIDAZOLE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Zhou, Q.J, Zhai, Y.J, Liu, M, Sun, F. | | Deposit date: | 2011-06-13 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex II via a water molecule mediated binding feature.
Protein Cell, 2, 2011
|
|
3KO1
 
 | | Cystal structure of thermosome from Acidianus tengchongensis strain S5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin | | Authors: | Huo, Y, Zhang, K, Hu, Z, Wang, L, Zhai, Y, Zhou, Q, Lander, G, He, Y, Zhu, J, Xu, W, Dong, Z, Sun, F. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of group II chaperonin in the open state.
Structure, 18, 2010
|
|
8KDE
 
 | | Cryo-EM structure of an intermediate-state complex during the process of photosystem II repair | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, A, Wang, Y, Liu, Z. | | Deposit date: | 2023-08-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for an early stage of the photosystem II repair cycle in Chlamydomonas reinhardtii.
Nat Commun, 15, 2024
|
|
5GUJ
 
 | |
6KLV
 
 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ANTIMYCIN, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6KLS
 
 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, Cytochrome c, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
