2RPT
 
 | |
2V7F
 
 | | Structure of P. abyssi RPS19 protein | | Descriptor: | CHLORIDE ION, RPS19E SSU RIBOSOMAL PROTEIN S19E | | Authors: | Gregory, L.A, Aguissa-Toure, A.H, Pinaud, N, Legrand, P, Gleizes, P.E, Fribourg, S. | | Deposit date: | 2007-07-30 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Basis of Diamond Blackfan Anemia: Structure and Function Analysis of Rps19.
Nucleic Acids Res., 35, 2007
|
|
2V94
 
 | |
2VOF
 
 | | Structure of mouse A1 bound to the Puma BH3-domain | | Descriptor: | BCL-2-BINDING COMPONENT 3, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VTY
 
 | | Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain swapped dimer | | Descriptor: | PROTEIN F1 | | Authors: | Kvansakul, M, Yang, H, Fairlie, W.D, Czabotar, P.E, Fischer, S.F, Perugini, M.A, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2008-05-19 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vaccinia Virus Anti-Apoptotic F1L is a Novel Bcl-2-Like Domain-Swapped Dimer that Binds a Highly Selective Subset of Bh3-Containing Death Ligands.
Cell Death Differ., 15, 2008
|
|
2VOG
 
 | | Structure of mouse A1 bound to the Bmf BH3-domain | | Descriptor: | BCL-2-MODIFYING FACTOR, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2G9P
 
 | | NMR structure of a novel antimicrobial peptide, latarcin 2a, from spider (Lachesana tarabaevi) venom | | Descriptor: | antimicrobial peptide Latarcin 2a | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2006-03-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Spatial structure and activity mechanism of a novel spider antimicrobial peptide.
Biochemistry, 45, 2006
|
|
6T1M
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1O
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6UNA
 
 | | Crystal structure of inactive p38gamma | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 12, SULFATE ION | | Authors: | Aoto, P.C, Stanfield, R.L, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-10-11 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A Dynamic Switch in Inactive p38 gamma Leads to an Excited State on the Pathway to an Active Kinase.
Biochemistry, 58, 2019
|
|
6UXO
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with DDM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V8U
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a modified Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*CP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A Conformational Switch in the Zinc Finger Protein Kaiso Mediates Differential Readout of Specific and Methylated DNA Sequences.
Biochemistry, 59, 2020
|
|
6UXQ
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with POPC and C8E4 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VBZ
 
 | | Crystal structure of the rat MLKL pseudokinase domain | | Descriptor: | MANGANESE (II) ION, Mixed lineage kinase domain-like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
6UXP
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylglycerol | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), Bcl-2 homologous antagonist/killer, GLYCEROL | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXN
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylserine | | Descriptor: | Bcl-2 homologous antagonist/killer, GLYCEROL, O-[(R)-{[(2R)-2,3-bis(octanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXM
 
 | |
6UXR
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with LysoPC | | Descriptor: | Bcl-2 homologous antagonist/killer, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1NU7
 
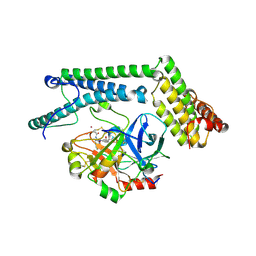 | | Staphylocoagulase-Thrombin Complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
1NO8
 
 | | SOLUTION STRUCTURE OF THE NUCLEAR FACTOR ALY RBD DOMAIN | | Descriptor: | ALY | | Authors: | Perez-Alvarado, G.C, Martinez-Yamout, M, Allen, M.M, Grosschedl, R, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-01-15 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Nuclear Factor ALY: Insights into Post-Transcriptional
Regulatory and mRNA Nuclear Export Processes
Biochemistry, 42, 2003
|
|
1NU9
 
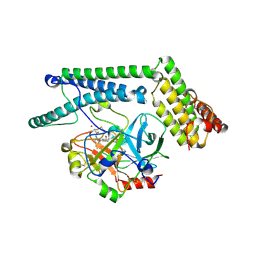 | | Staphylocoagulase-Prethrombin-2 complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
4YGW
 
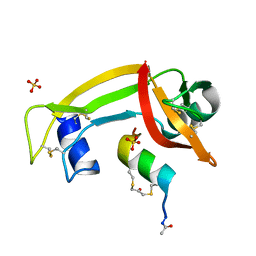 | | RNase S in complex with stabilized S peptide | | Descriptor: | 1-hydroxypropan-2-one, Ribonuclease A C2, S-peptide: ACE-LYS-GLU-THR-ALA-ALA-HCS-LYS-PHE-GLU-HCS-GLN-HIS-MET-ASP-SER, ... | | Authors: | Assem, N, Ferreira, D, Wolan, D.W, Dawson, P.E. | | Deposit date: | 2015-02-26 | | Release date: | 2015-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Acetone-Linked Peptides: A Convergent Approach for Peptide Macrocyclization and Labeling.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4Z6I
 
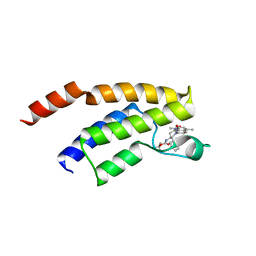 | | Crystal structure of BRD9 bromodomain in complex with a substituted valerolactam quinolone ligand | | Descriptor: | Bromodomain-containing protein 9, tert-butyl [(2R,3S)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxo-2-phenylpiperidin-3-yl]carbamate | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4ZIH
 
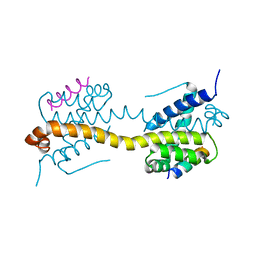 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
