7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCG
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCK
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
6OGN
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SGC8158 chemical probe | | Descriptor: | 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, Protein arginine N-methyltransferase 7, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response.
Nat Commun, 11, 2020
|
|
8CTH
 
 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Phe) complex | | Descriptor: | Phe-tRNA, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase, ... | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fishcer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
8CTI
 
 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Val) complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4, tRNA-Val-TAC-2-1 | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fischer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
7N40
 
 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | Descriptor: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | Authors: | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
8J2P
 
 | | Crystal structure of PML B-box2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
8J25
 
 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
7U20
 
 | | Crystal structure of human METTL1 and WDR4 complex | | Descriptor: | SULFATE ION, tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Li, J, Nowak, R.P, Fischer, E.S, Gregory, R. | | Deposit date: | 2022-02-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
4Y5Y
 
 | | Diabody 330 complex with EpoR | | Descriptor: | Erythropoietin receptor, GLYCEROL, diabody 330 VH domain, ... | | Authors: | Moraga, I, Guo, F, Ozkan, E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-02-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands.
Cell, 160, 2015
|
|
4Y5X
 
 | | Diabody 305 complex with EpoR | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, Erythropoietin receptor, ... | | Authors: | Moraga, I, Guo, F, Ozkan, E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-02-12 | | Release date: | 2015-03-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands.
Cell, 160, 2015
|
|
4Y5V
 
 | | Diabody 305 complex with EpoR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Diabody 305 VL domain, Erythropoietin receptor, ... | | Authors: | Moraga, I, Guo, F, Ozkan, E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands.
Cell, 160, 2015
|
|
5H7U
 
 | | NMR structure of eIF3 36-163 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit C | | Authors: | Nagata, T, Obayashi, E. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5.
Cell Rep, 18, 2017
|
|
8CAP
 
 | |
8CAL
 
 | |
8CB0
 
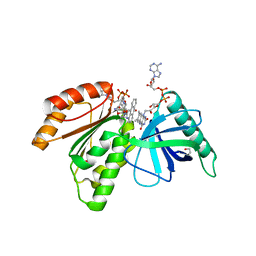 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with M41 and NADP+ | | Descriptor: | 1,2-ETHANEDIOL, 15-(1,4-dioxa-8-azaspiro[4.5]decan-8-yl)-14-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(16),2(7),3,5,9,11,13(17),14-octaen-8-one, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
8CAO
 
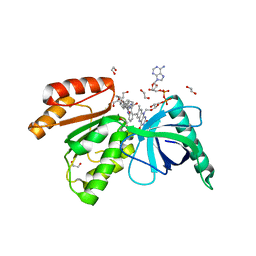 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with CA24 | | Descriptor: | 1,2-ETHANEDIOL, 8-[2-(4-cyclohexylphenyl)quinolin-4-yl]carbonyl-1,3,8-triazaspiro[4.5]decane-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
8CAK
 
 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with M41 | | Descriptor: | 1,2-ETHANEDIOL, 15-(1,4-dioxa-8-azaspiro[4.5]decan-8-yl)-14-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(16),2(7),3,5,9,11,13(17),14-octaen-8-one, DIMETHYL SULFOXIDE, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
5B23
 
 | |
6JOD
 
 | | Angiotensin II type 2 receptor with ligand | | Descriptor: | Angiotensin II, Heavy chain of 4A03Fab, Light chain of 4A03Fab, ... | | Authors: | Asada, H, Iwata, S, Hirata, K, Shimamura, T. | | Deposit date: | 2019-03-20 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of Angiotensin II Type 2 Receptor with Endogenous Peptide Hormone.
Structure, 28, 2020
|
|
7D6P
 
 | |
7D6T
 
 | |
