8J6S
 
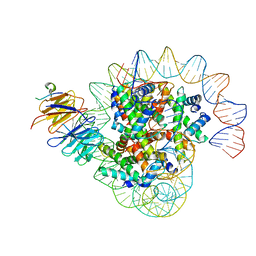 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8J6T
 
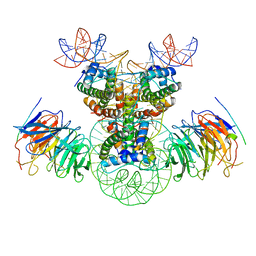 | | Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQF
 
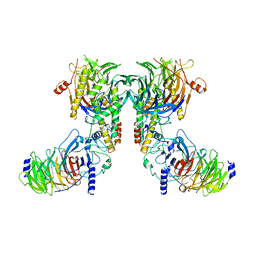 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQG
 
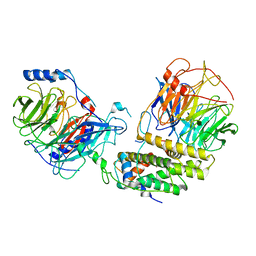 | | Cryo-EM structure of the monomeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
4KQL
 
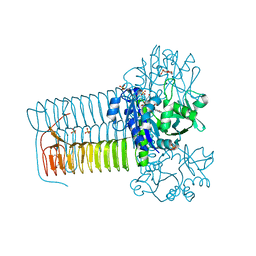 | | Hin GlmU bound to WG578 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-(4-{[3-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-5-methoxybenzoyl]amino}phenyl)pyridine-2-carboxamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
6ADM
 
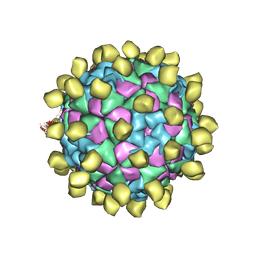 | |
6ADR
 
 | |
6ADL
 
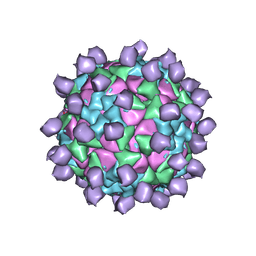 | |
5GNU
 
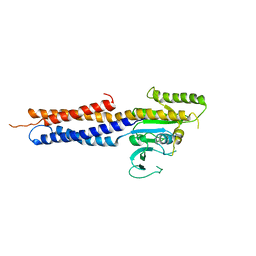 | | the structure of mini-MFN1 apo | | Descriptor: | Mitofusin-1 | | Authors: | Yan, L, Yu, C, Ming, Z, Lou, Z, Rao, Z, Lou, J. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.113 Å) | | Cite: | BDLP-like folding of Mitofusin 1
To Be Published
|
|
6ADS
 
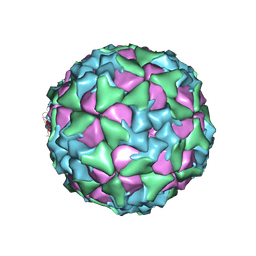 | | Structure of Seneca Valley Virus in acidic conditions | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6KRX
 
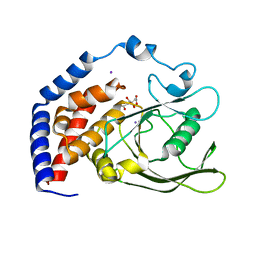 | | Crystal Structure of AtPTP1 at 1.7 angstrom | | Descriptor: | CITRATE ANION, IODIDE ION, Protein-tyrosine-phosphatase PTP1 | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | AtPTP1 positively mediates brassinosteroid signaling from receptor kinases to GSK3-like kinase BIN2
To Be Published
|
|
6ADT
 
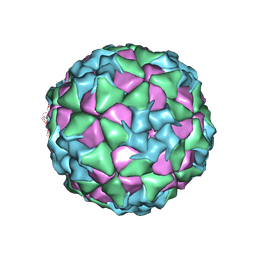 | | Structure of Seneca Valley Virus in neutral condition | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6KRW
 
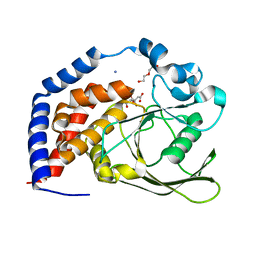 | | Crystal Structure of AtPTP1 at 1.4 angstrom | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of AtPTP1 at 1.4 Angstroms
To Be Published
|
|
6LW2
 
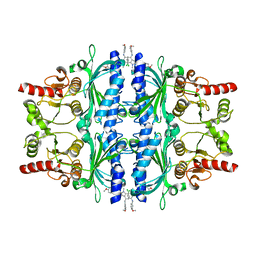 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 7-chloranyl-4-[(3-methoxyphenyl)amino]-N-(4-methoxyphenyl)sulfonyl-1-methyl-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of N -Arylsulfonyl-Indole-2-Carboxamide Derivatives as Potent, Selective, and Orally Bioavailable Fructose-1,6-Bisphosphatase Inhibitors-Design, Synthesis, In Vivo Glucose Lowering Effects, and X-ray Crystal Complex Analysis.
J.Med.Chem., 63, 2020
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
6JSJ
 
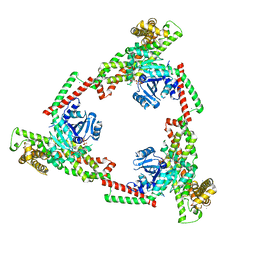 | |
6U2D
 
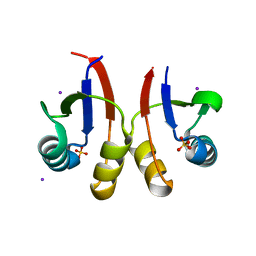 | | PmtCD peptide exporter basket domain | | Descriptor: | ABC transporter ATP-binding protein, IODIDE ION, SULFATE ION | | Authors: | Zeytuni, N, Strynadka, N.C.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
7RYJ
 
 | |
4R8M
 
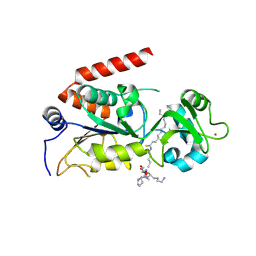 | | Human SIRT2 crystal structure in complex with BHJH-TM1 | | Descriptor: | BHJH-TM1 peptide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Teng, Y.B, Hao, Q, Lin, H.N, Jing, H. | | Deposit date: | 2014-09-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Efficient Demyristoylase Activity of SIRT2 Revealed by Kinetic and Structural Studies
Sci Rep, 5, 2015
|
|
7JKB
 
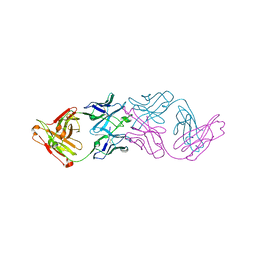 | | 2xVH Fab | | Descriptor: | Anti-Her2, Anti-lysozyme | | Authors: | Lord, D.M, Zhou, Y.F. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Bringing the Heavy Chain to Light: Creating a Symmetric, Bivalent IgG-Like Bispecific.
Antibodies, 9, 2020
|
|
8ZMN
 
 | | Crystal structure of ANTXR1 | | Descriptor: | Anthrax toxin receptor 1, SODIUM ION | | Authors: | Zheng, H, Hu, J, Fu, L, Chen, R. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray structure of Anthrax toxin receptor 1 APO from Rattus norvegicus
To Be Published
|
|
7E2R
 
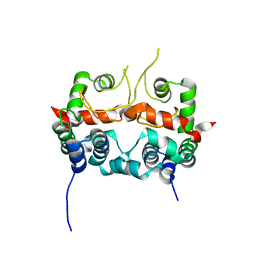 | | The ligand-free structure of Arabidopsis thaliana GUN4 | | Descriptor: | Tetrapyrrole-binding protein, chloroplastic | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
5BNR
 
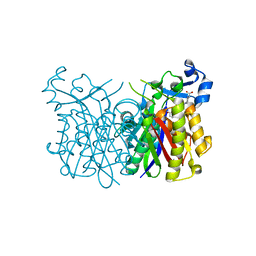 | | E. coli Fabh with small molecule inhibitor 2 | | Descriptor: | 1-{5-[2-fluoro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5BQS
 
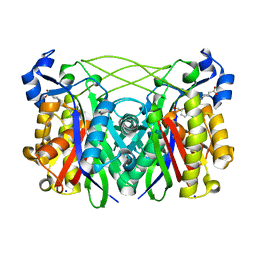 | | S. Pneumoniae Fabh with small molecule inhibitor 4 | | Descriptor: | 1-{5-[2-chloro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SODIUM ION | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-29 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
4XWW
 
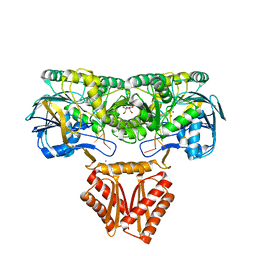 | | Crystal structure of RNase J complexed with RNA | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
