2RIS
 
 | |
6TQK
 
 | | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
2RIU
 
 | | Alternative models for two crystal structures of Candida albicans 3,4-dihydroxy-2-butanone 4-phosphate synthase- alternate interpreation | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE | | Authors: | Stenkamp, R.E, Le Trong, I. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternative models for two crystal structures of Candida albicans 3,4-dihydroxy-2-butanone 4-phosphate synthase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4GK7
 
 | | yeast 20S proteasome in complex with the Syringolin-Glidobactin chimera | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Groll, M, Stein, M.L, Bachmann, A. | | Deposit date: | 2012-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activity enhancement of the synthetic syrbactin proteasome inhibitor hybrid and biological evaluation in tumor cells.
Biochemistry, 51, 2012
|
|
6TQL
 
 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
4ZVZ
 
 | | Co-crystal structures of PP5 in complex with 5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid | | Descriptor: | (1R,2S,3R,4S,5S)-5-(propoxymethyl)-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Wierzbicki, A, Honkanen, R.E. | | Deposit date: | 2015-05-18 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C.
Biochem. Pharmacol., 109, 2016
|
|
6TRM
 
 | | Solution structure of the antifungal protein PAFC | | Descriptor: | Pc21g12970 protein | | Authors: | Czajlik, A, Holzknecht, J, Marx, F, Batta, G. | | Deposit date: | 2019-12-19 | | Release date: | 2020-10-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics, and New Antifungal Aspects of the Cysteine-Rich Miniprotein PAFC.
Int J Mol Sci, 22, 2021
|
|
6R45
 
 | | Crystal structure of eukaryotic O-GlcNAcase HAT-like domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, O-GlcNAcase | | Authors: | Raimi, O.G, Gorelik, A, Hopkins-Navratilova, I, Aristotelous, T, Ferenbach, A, vanAalten, D. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Crystal structure of eukaryotic O-GlcNAcase HAT-like domain
To Be Published
|
|
4ZX2
 
 | | Co-crystal structures of PP5 in complex with 5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid | | Descriptor: | (1S,2R,3S,4R,5S)-5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Wierzbicki, A, Honkanen, R.E. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C.
Biochem. Pharmacol., 109, 2016
|
|
5K59
 
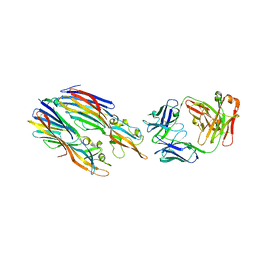 | | Crystal structure of LukGH from Staphylococcus aureus in complex with a neutralising antibody | | Descriptor: | CHLORIDE ION, Fab heavy chain, Fab light chain, ... | | Authors: | Welin, M, Logan, D.T, Badarau, A, Mirkina, I, Zauner, G, Dolezilkova, I, Nagy, E. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies.
Mabs, 8, 2016
|
|
6YN9
 
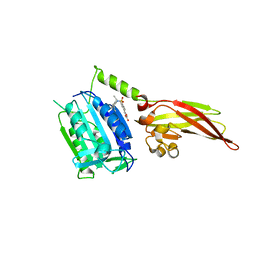 | | MALT1(329-728) in complex with a sulfonamide containing compound | | Descriptor: | 5-[4-[(2,6-diethylphenyl)sulfamoyl]-3-methyl-phenyl]furan-3-carboxylic acid, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
5LN2
 
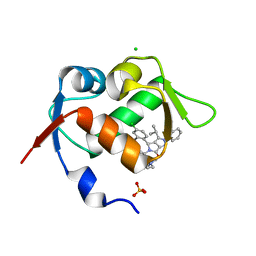 | |
6Z10
 
 | | Crystal structure of a humanized (K18E, K269N) rat succinate receptor SUCNR1 (GPR91) in complex with a nanobody and antagonist | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[[3-[4-chloranyl-2-fluoranyl-5-[(3~{R})-piperidin-3-yl]oxy-phenyl]-2-fluoranyl-phenyl]carbonylamino]-5-fluoranyl-phenyl]ethanoic acid, CHLORIDE ION, ... | | Authors: | Haffke, M, Villard, F. | | Deposit date: | 2020-05-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Discovery and Optimization of Novel SUCNR1 Inhibitors: Design of Zwitterionic Derivatives with a Salt Bridge for the Improvement of Oral Exposure.
J.Med.Chem., 63, 2020
|
|
6YN8
 
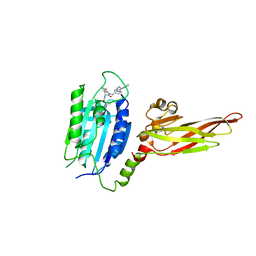 | | Human MALT1(334-719) in complex with a tetrazole containing compound | | Descriptor: | 3-azanyl-3-methyl-~{N}-[(3~{R})-4-oxidanylidene-5-[[4-[2-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]-2,3-dihydro-1,5-benzoxazepin-3-yl]butanamide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
6GGN
 
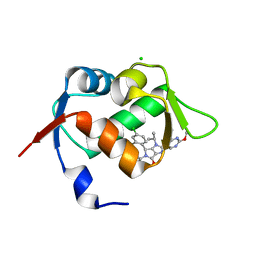 | | In vitro and in vivo characterization of a novel, highly potent p53-MDM2 inhibitor | | Descriptor: | (4~{S})-4-(4-chloranyl-2-methyl-phenyl)-5-(5-chloranyl-2-methyl-phenyl)-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Kallen, J. | | Deposit date: | 2018-05-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro and in vivo characterization of a novel, highly potent p53-MDM2 inhibitor.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6VW1
 
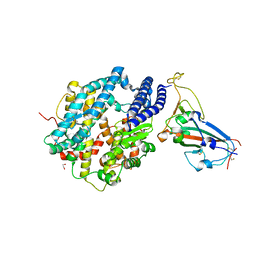 | | Structure of SARS-CoV-2 chimeric receptor-binding domain complexed with its receptor human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Ye, G, Shi, K, Wan, Y.S, Aihara, H, Li, F. | | Deposit date: | 2020-02-18 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of receptor recognition by SARS-CoV-2.
Nature, 581, 2020
|
|
1RD5
 
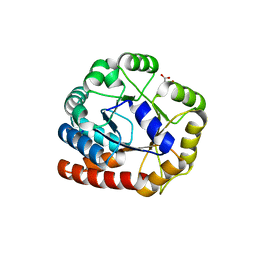 | | Crystal structure of Tryptophan synthase alpha chain homolog BX1: a member of the chemical plant defense system | | Descriptor: | MALONIC ACID, Tryptophan synthase alpha chain, chloroplast | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2003-11-05 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
2V2V
 
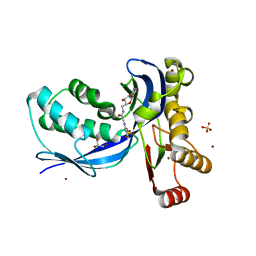 | | IspE in complex with ligand | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, 5'-[(1H-BENZIMIDAZOL-2-YLACETYL)AMINO]-5'-DEOXYCYTIDINE, BROMIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Characterization of Cytidine Derivatives that Inhibit the Kinase Ispe of the Non-Mevalonate Pathway for Isoprenoid Biosynthesis.
Chemmedchem, 3, 2008
|
|
2V2Q
 
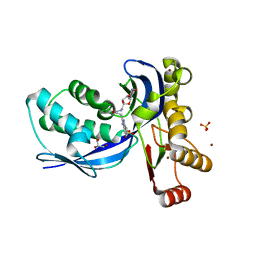 | | IspE in complex with ligand | | Descriptor: | 4-AMINO-1-(5-{[3-(1H-BENZIMIDAZOL-2-YL)PROPANOYL]AMINO}-5-DEOXY-ALPHA-L-LYXOFURANOSYL)PYRIMIDIN-2(1H)-ONE, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL KINASE, BROMIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Characterization of Cytidine Derivatives that Inhibit the Kinase Ispe of the Non-Mevalonate Pathway for Isoprenoid Biosynthesis.
Chemmedchem, 3, 2008
|
|
2MD8
 
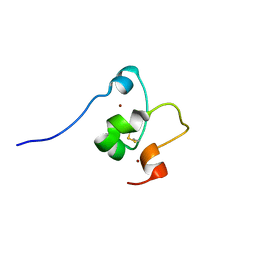 | |
2MD7
 
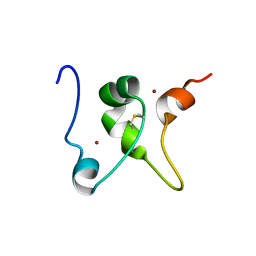 | |
6D92
 
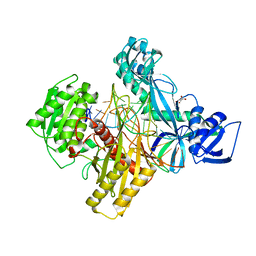 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-A non-canonical pair at position 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-27 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D8F
 
 | | RsAgo Ternary Complex with Guide RNA and Target DNA Containing T-T Bulge Within the Seed Segment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*TP*TP*TP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D8A
 
 | | RsAgo Ternary Complex with guide RNA and Target DNA Containing A-A Bulge Within the Seed Segment of the Target Strand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D8P
 
 | | Ternary RsAgo Complex Containing Guide RNA Paired with Target DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CACODYLATE ION, ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
