4Q5W
 
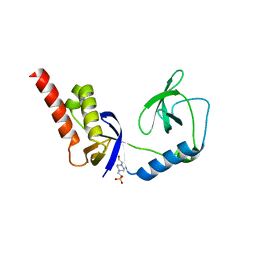 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
8U4K
 
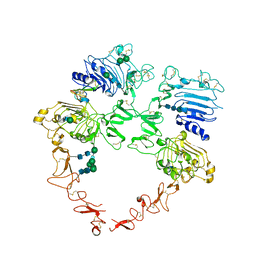 | | Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4I
 
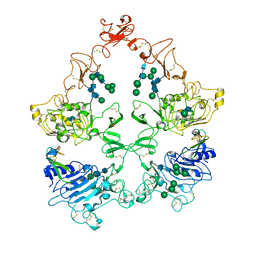 | | Structure of the HER4/NRG1b Homodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4L
 
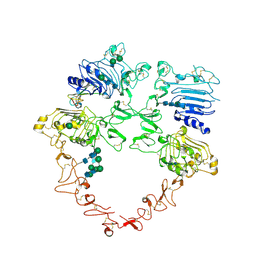 | | Structure of the HER2/HER4/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4J
 
 | | Structure of the HER4/BTC Homodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8FHC
 
 | | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium | | Descriptor: | BROMIDE ION, CHOLIC ACID, FE (III) ION, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium
To Be Published
|
|
8FH5
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, Aldo-keto reductase family 1 member B1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001
To Be Published
|
|
8FH6
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001
To Be Published
|
|
8FH9
 
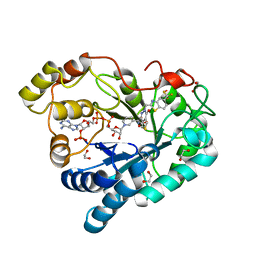 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007 | | Descriptor: | (4-oxo-3-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-3,4-dihydrothieno[3,4-d]pyridazin-1-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007
To Be Published
|
|
8FHB
 
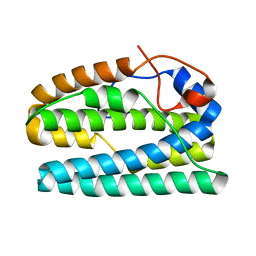 | |
8FH7
 
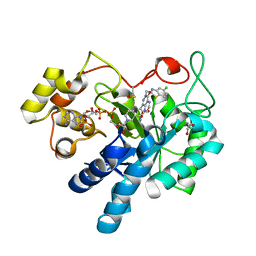 | |
8FIF
 
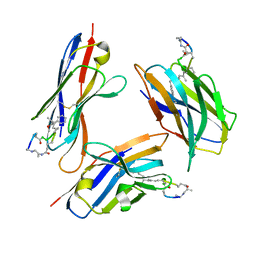 | |
8FH8
 
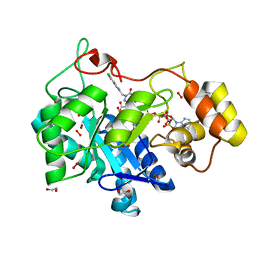 | |
5KJX
 
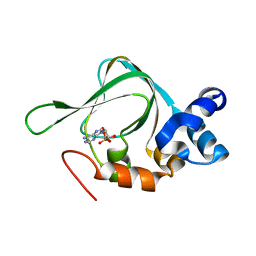 | | Co-crystal Structure of PKA RI alpha CNB-B domain with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
5KJY
 
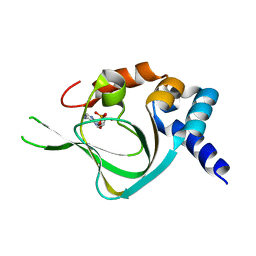 | | Co-crystal structure of PKA RI alpha CNB-B mutant (G316R/A336T) with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
5KJZ
 
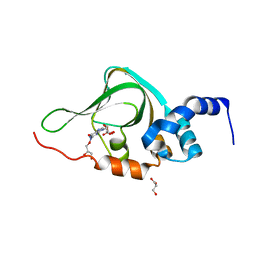 | | Co-crystal structure of PKA RI alpha CNB-B mutant (G316R/A336T) with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
5UBK
 
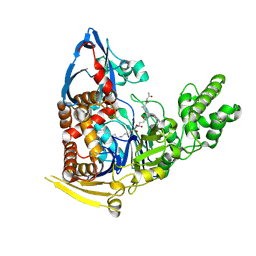 | | Inactive S1A/N269D-cpPvdQ mutant in complex with the pyoverdine precursor PVDIq reveals a specific binding pocket for the D-Tyr of this substrate | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, N-[(1R)-1-{(6S)-6-[(2-amino-2-oxoethyl)carbamoyl]-1,4,5,6-tetrahydropyrimidin-2-yl}-2-(4-hydroxyphenyl)ethyl]-N~2~-tetradecanoyl-L-glutamine | | Authors: | Mascarenhas, R, Catlin, D, Wu, R, Clevenger, K, Fast, W, Liu, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-01 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Circular Permutation Reveals a Chromophore Precursor Binding Pocket of the Siderophore Tailoring Enzyme PvdQ
To Be Published
|
|
5VWR
 
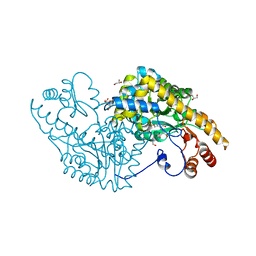 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP)-alpha-ketoglutarate | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, Aspartate aminotransferase, GLYCEROL | | Authors: | Mascarenhas, R, Liu, D, Le, H, Silverman, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
5VWO
 
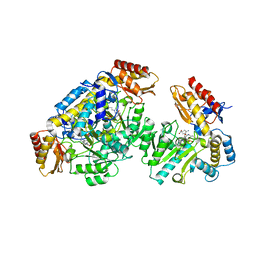 | | Ornithine aminotransferase inactivated by (1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP) | | Descriptor: | (1S,3S,4E)-3-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-4-iminocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Mascarenhas, R, Liu, D, Le, H, Silverman, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
4ZO3
 
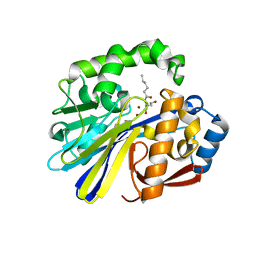 | | AidC, a Dizinc Quorum-Quenching Lactonase, in complex with a product N-hexnoyl-L-homoserine | | Descriptor: | Acylhomoserine lactonase, N-hexanoyl-L-homoserine, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
4ZO2
 
 | | AidC, a Dizinc Quorum-Quenching Lactonase | | Descriptor: | Acylhomoserine lactonase, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
5VWQ
 
 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase | | Authors: | Mascarenhas, R, Lehrer, H, Liu, D, Ringe, D. | | Deposit date: | 2017-05-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
4FSC
 
 | | Crystal Structure of Bacillus thuringiensis PlcR in its apo form | | Descriptor: | Transcriptional activator PlcR protein | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2012-06-27 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4NEJ
 
 | | Small molecular fragment bound to crystal contact interface of Interleukin-2 | | Descriptor: | 5-methylfuran-2-carboxylic acid, Interleukin-2 | | Authors: | Brenke, R, Jehle, S, Vajda, S, Allen, K.N, Kozakov, D. | | Deposit date: | 2013-10-29 | | Release date: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Small molecular fragments bound to binding energy hot-spot in crystal contact interface of Interleukin-2
To be Published
|
|
3U3W
 
 | | Crystal Structure of Bacillus thuringiensis PlcR in complex with the peptide PapR7 and DNA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*AP*AP*AP*TP*AP*TP*TP*GP*CP*AP*TP*AP*G)-3', 5'-D(P*CP*TP*AP*TP*GP*CP*AP*AP*TP*AP*TP*TP*TP*CP*AP*TP*AP*T)-3', C-terminus heptapeptide from PapR protein, ... | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2011-10-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
