1XAM
 
 | | Cobalt hexammine induced tautameric shift in Z-DNA: structure of d(CGCGCA).d(TGCGCG) in two crystal forms. | | Descriptor: | CGCGCA, COBALT HEXAMMINE(III), TG, ... | | Authors: | Thiyagarajan, S, Rajan, S.S, Gautham, N. | | Deposit date: | 2004-08-26 | | Release date: | 2004-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cobalt hexammine induced tautomeric shift in Z-DNA: the structure of d(CGCGCA)*d(TGCGCG) in two crystal forms.
Nucleic Acids Res., 32, 2004
|
|
1XA2
 
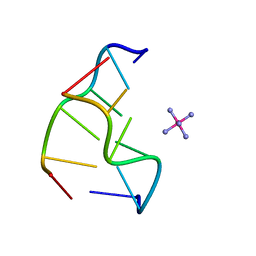 | |
1LJX
 
 | | THE STRUCTURE OF D(TPGPCPGPCPA)2 AT 293K: COMPARISON OF THE EFFECT OF SEQUENCE AND TEMPERATURE | | Descriptor: | 5'-D(*TP*GP*CP*GP*CP*A)-3', MAGNESIUM ION | | Authors: | Thiyagarajan, S, Satheesh Kumar, P, Rajan, S.S, Gautham, N. | | Deposit date: | 2002-04-23 | | Release date: | 2002-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of d(TGCGCA)2 at 293 K: comparison of the effects of sequence and temperature.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1T1B
 
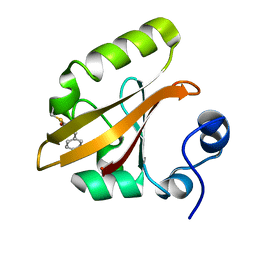 | | Late intermediate IL2 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T1C
 
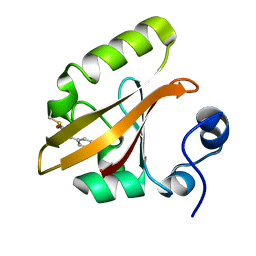 | | Late intermediate IL3 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T18
 
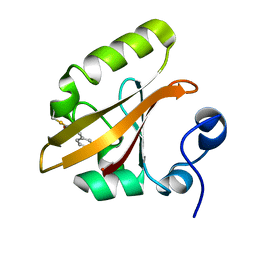 | | Early intermediate IE1 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T19
 
 | | Early intermediate IE2 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T1A
 
 | | Late intermediate IL1 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
3FK5
 
 | |
1MZU
 
 | |
3JRU
 
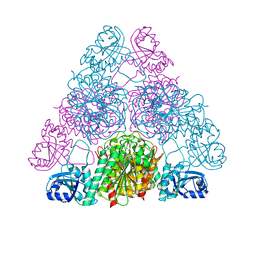 | | Crystal structure of Leucyl Aminopeptidase (pepA) from Xoo0834,Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | CALCIUM ION, CARBONATE ION, Probable cytosol aminopeptidase, ... | | Authors: | Natarajan, S, Huynh, K.-H, Kang, L.W. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Leucyl Aminopeptidase (pepA) from Xoo0834,Xanthomonas oryzae pv. oryzae KACC10331
to be published
|
|
3K89
 
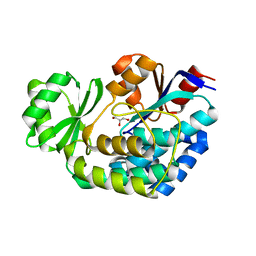 | |
3NX6
 
 | |
3R97
 
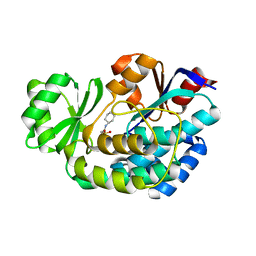 | | Crystal structure of malonyl-CoA:acyl carrier protein transacylase (FabD), Xoo0880, from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Malonyl CoA-ACP transacylase | | Authors: | Natarajan, S, Jung, J.W, Kang, L.W. | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of malonyl-CoA:acyl carrier protein transacylase (FabD), Xoo0880, from Xanthomonas oryzae pv. oryzae KACC10331
To be Published
|
|
3KET
 
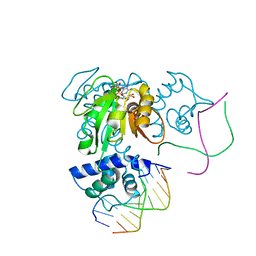 | | Crystal structure of a Rex-family transcriptional regulatory protein from Streptococcus agalactiae bound to a palindromic operator | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*TP*CP*AP*CP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Thiyagarajan, S, Logan, D, von Wachenfeldt, C. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence.
Plos Pathog., 17, 2021
|
|
3KEO
 
 | | Crystal Structure of a Rex-family transcriptional regulatory protein from Streptococcus agalactiae complexed with NAD+ | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thiyagarajan, S, Logan, D, von Wachenfeldt, C. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence.
Plos Pathog., 17, 2021
|
|
3KEQ
 
 | |
6H60
 
 | | pseudo-atomic structural model of the E3BP component of the human pyruvate dehydrogenase multienzyme complex | | Descriptor: | Pyruvate dehydrogenase protein X component, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
6H55
 
 | | core of the human pyruvate dehydrogenase (E2) | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
4JLL
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1 covalently bound with FP-alkyne, Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JVV
 
 | | Crystal structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1, covalently bound with diisopropyl fluorophosphate (DFP), Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | evolved variant of the computationally designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JCA
 
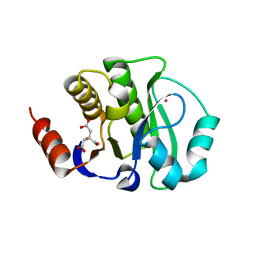 | | Crystal Structure of the apo form of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1. Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | CITRIC ACID, RUBIDIUM ION, serine hydrolase | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4DRT
 
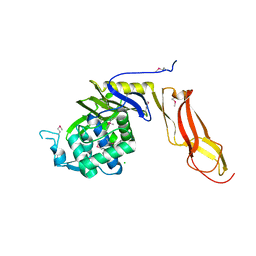 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4F2V
 
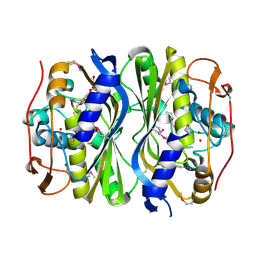 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3V45
 
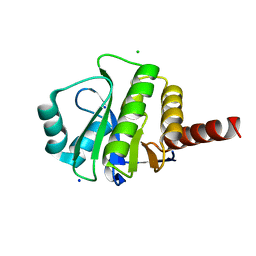 | | Crystal Structure of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium Target OR130 | | Descriptor: | CHLORIDE ION, SODIUM ION, Serine hydrolase OSH55 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
