2HQI
 
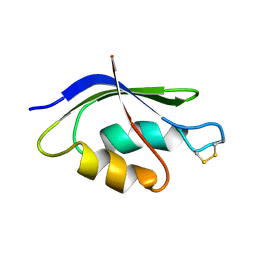 | | NMR SOLUTION STRUCTURE OF THE OXIDIZED FORM OF MERP, 14 STRUCTURES | | Descriptor: | MERCURIC TRANSPORT PROTEIN | | Authors: | Qian, H, Sahlman, L, Eriksson, P.O, Hambreus, C, Edlund, U, Sethson, I. | | Deposit date: | 1998-03-31 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the oxidized form of MerP, a mercuric ion binding protein involved in bacterial mercuric ion resistance.
Biochemistry, 37, 1998
|
|
1HQI
 
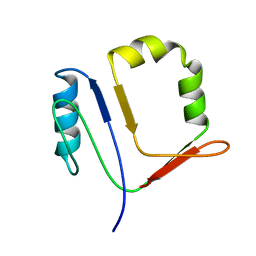 | |
6VP0
 
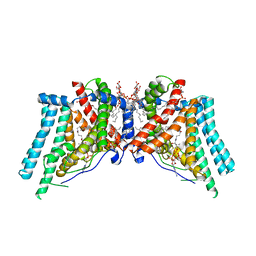 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
1JOY
 
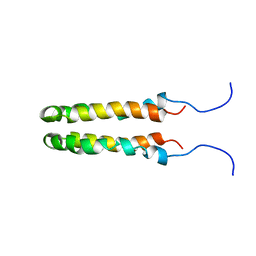 | | SOLUTION STRUCTURE OF THE HOMODIMERIC DOMAIN OF ENVZ FROM ESCHERICHIA COLI BY MULTI-DIMENSIONAL NMR. | | Descriptor: | PROTEIN (ENVZ_ECOLI) | | Authors: | Tomomori, C, Tanaka, T, Dutta, R, Park, H, Saha, S.K, Zhu, Y, Ishima, R, Liu, D, Tong, K.I, Kurokawa, H, Qian, H, Inouye, M, Ikura, M. | | Deposit date: | 1998-12-28 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ.
Nat.Struct.Biol., 6, 1999
|
|
3T5T
 
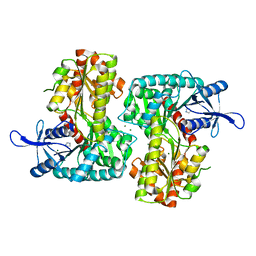 | |
3T7D
 
 | | Vall from streptomyces hygroscopicus in complex with trehalose | | Descriptor: | IMIDAZOLE, MAGNESIUM ION, Putative glycosyltransferase, ... | | Authors: | Zhang, H, Zheng, L, Qian, H, Chen, J. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the substrate specificity of ValL
To be Published
|
|
4ILE
 
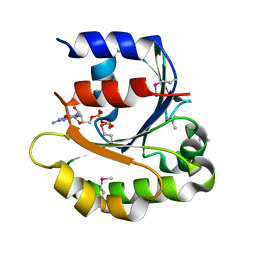 | |
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
4A3N
 
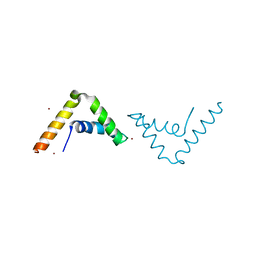 | | Crystal Structure of HMG-BOX of Human SOX17 | | Descriptor: | TRANSCRIPTION FACTOR SOX-17, ZINC ION | | Authors: | Gao, N, Gao, H, Qian, H, Si, S, Xie, Y. | | Deposit date: | 2011-10-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Human Transcription Factor Sry-Related Box 17 Binding to DNA.
Protein Pept.Lett., 20, 2013
|
|
7ETW
 
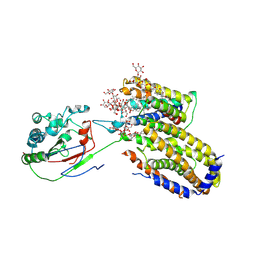 | | Cryo-EM structure of Scap/Insig complex in the present of digitonin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Digitonin, Insulin-induced gene 2 protein, ... | | Authors: | Yan, R, Cao, P, Song, W, Li, Y, Wang, T, Qian, H, Yan, C, Yan, N. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for sterol sensing by Scap and Insig
Cell Rep, 35, 2021
|
|
6P2J
 
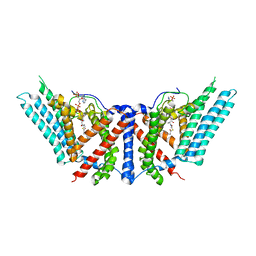 | | Dimeric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
6P2P
 
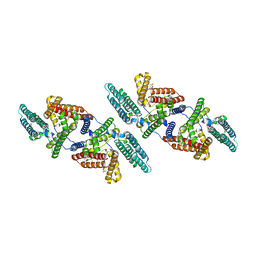 | | Tetrameric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
6W5V
 
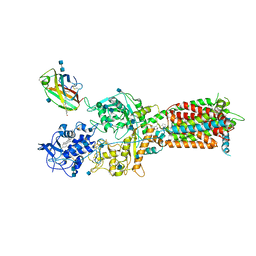 | | NPC1-NPC2 complex structure at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5R
 
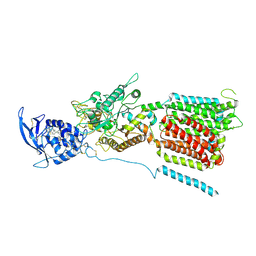 | | NPC1 structure in Nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5S
 
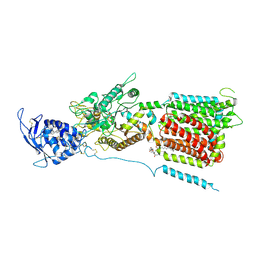 | | NPC1 structure in GDN micelles at pH 8.0 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5U
 
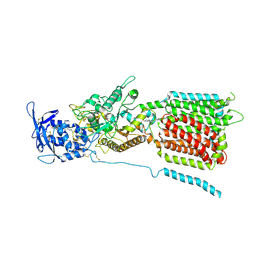 | | NPC1 structure in GDN micelles at pH 5.5, conformation b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5T
 
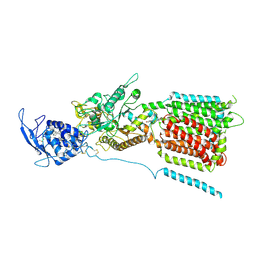 | | NPC1 structure in GDN micelles at pH 5.5, conformation a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6N7G
 
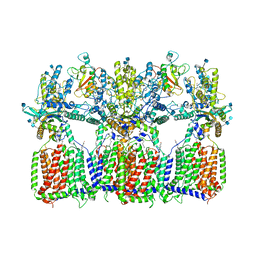 | | Cryo-EM structure of tetrameric Ptch1 in complex with ShhNp (form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.
Nat Commun, 10, 2019
|
|
6N7K
 
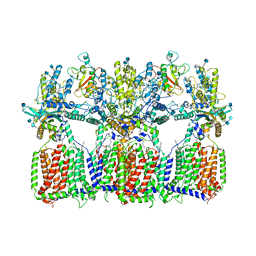 | | Cryo-EM structure of tetrameric Ptch1 in complex with ShhNp (form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.
Nat Commun, 10, 2019
|
|
6N7H
 
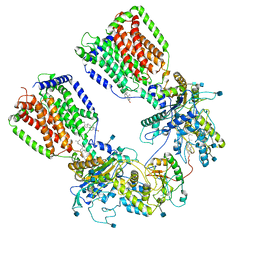 | | Cryo-EM structure of the 2:1 hPtch1-Shhp complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.
Nat Commun, 10, 2019
|
|
5XJY
 
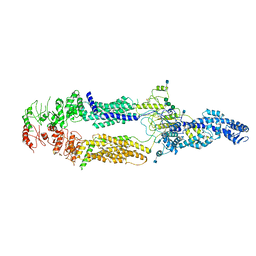 | | Cryo-EM structure of human ABCA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette sub-family A member 1, ... | | Authors: | Qian, H.W, Yan, N, Gong, X. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the Human Lipid Exporter ABCA1.
Cell, 169, 2017
|
|
5XUN
 
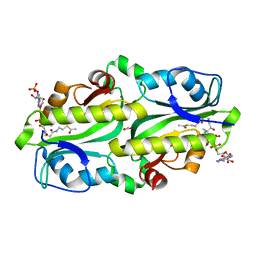 | | Crystal structure of Y145F mutant of KacT | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, Acetyltransferase | | Authors: | Qian, H.L, Yao, Q.Q, Gan, J.H, Ou, H.Y. | | Deposit date: | 2017-06-23 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of acetyltransferase-type toxin-antitoxin locus in Klebsiella pneumoniae
Mol. Microbiol., 108, 2018
|
|
6DS5
 
 | | Cryo EM structure of human SEIPIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Seipin | | Authors: | Yan, R.H, Qian, H.W, Yan, N, Yang, H.Y. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human SEIPIN Binds Anionic Phospholipids.
Dev. Cell, 47, 2018
|
|
6DMB
 
 | | Cryo-EM structure of human Ptch1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-06-04 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the recognition of Sonic Hedgehog by human Patched1.
Science, 361, 2018
|
|
6DMO
 
 | | Cryo-EM structure of human Ptch1 with three mutations L282Q/T500F/P504L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-06-05 | | Release date: | 2018-07-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the recognition of Sonic Hedgehog by human Patched1.
Science, 361, 2018
|
|
